For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTNVKDDKTLRKFRRERALGRYLLNPAVKGMGKLGLRTALATELETIGRKSGQARRVPVSAQFDANGAWI ICQHGTRSGWGRNIVDNPNIRIRQGTRWRTGVATLRPDDDVVARGRKFGRLGATVVKALETTPVSVRIDF TD
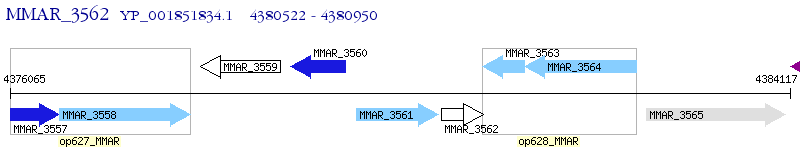
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3562 | - | - | 100% (142) | hypothetical protein MMAR_3562 |
| M. marinum M | MMAR_2750 | - | 1e-18 | 43.88% (98) | hypothetical protein MMAR_2750 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1902c | - | 1e-18 | 44.90% (98) | hypothetical protein Mb1902c |
| M. gilvum PYR-GCK | Mflv_4462 | - | 4e-15 | 36.89% (103) | hypothetical protein Mflv_4462 |
| M. tuberculosis H37Rv | Rv1871c | - | 1e-18 | 44.90% (98) | hypothetical protein Rv1871c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4636 | - | 8e-62 | 79.41% (136) | hypothetical protein MAB_4636 |
| M. avium 104 | MAV_2845 | - | 5e-19 | 43.88% (98) | hypothetical protein MAV_2845 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2795 | - | 6e-77 | 97.18% (142) | hypothetical protein MUL_2795 |
| M. vanbaalenii PYR-1 | Mvan_1899 | - | 8e-16 | 35.16% (128) | hypothetical protein Mvan_1899 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1902c|M.bovis_AF2122/97 ----------------MNAAMNLKREFVHRVQRFVVN-------------
Rv1871c|M.tuberculosis_H37Rv ----------------MNAAMNLKREFVHRVQRFVVN-------------
MAV_2845|M.avium_104 --------------------MSLKRRVVHRVQRLLVN-------------
Mflv_4462|M.gilvum_PYR-GCK ------------------MAETTRDRVTKFFQKNIAN-------------
Mvan_1899|M.vanbaalenii_PYR-1 -----------------MTADTLRDRVTKFFQKNVAN-------------
MMAR_3562|M.marinum_M ------------------MTNVKDDKTLRKFRRERALGRYLLNPAVKGMG
MUL_2795|M.ulcerans_Agy99 ------------------MTNVKDDKTLRKFRRERALGRYLLNPAVKGMG
MAB_4636|M.abscessus_ATCC_1997 MDHDPAAPQEQRRRSIPKKEKFMTDKTLRKFRRERAMGRYLLNPAVGALA
. : .:: .
Mb1902c|M.bovis_AF2122/97 PIGRQLP-MTMLETIGRKTGQPRRTAVGGRVVDNQFWMVSEHGEHSDYVY
Rv1871c|M.tuberculosis_H37Rv PIGRQLP-MTMLETIGRKTGQPRRTAVGGRVVDNQFWMVSEHGEHSDYVY
MAV_2845|M.avium_104 PVGRQLP-MTMLETTGRKSGKPRRTAVGGRVVDNQFWMVSEHGEHSDYVR
Mflv_4462|M.gilvum_PYR-GCK RVMRHIPIQTLLETTGRKSGLPRTTPLGGRRIGDEFWFVSEFGDRSQYVR
Mvan_1899|M.vanbaalenii_PYR-1 RVMRQMPFQTLLETTGRKSGQPRTTPLGGRRIGDAFWFVSEFGDRSQYIR
MMAR_3562|M.marinum_M KLGLRTALATELETIGRKSGQARRVPVSAQFDANGAWIICQHGTRSGWGR
MUL_2795|M.ulcerans_Agy99 KLGLRTALATGLETIGRKTGQARRVPVSAQFDANGAWIICQHGTRSGWGR
MAB_4636|M.abscessus_ATCC_1997 KLGVRTALASELETTGRKTGQLRRVPVSVQFDSTGAWVICQHGTRSGWGS
: : . : *** ***:* * ..:. : *.:.:.* :* :
Mb1902c|M.bovis_AF2122/97 NIKANPAVRVRIGGRWRSGTAYLLPDDDPRQRLRGLPRLNSAGVRAMGTD
Rv1871c|M.tuberculosis_H37Rv NIKANPAVRVRIGGRWRSGTAYLLPDDDPRQRLRGLPRLNSAGVRAMGTD
MAV_2845|M.avium_104 NIKANPAVRVRVGGRWRTGTAHLLPDDDPVQRLGNLPRLNSAMVRLMGSD
Mflv_4462|M.gilvum_PYR-GCK NIMADQNVRLRLRGRWHRGIAHLLPEDDAQARMRMLPRYNNVGVRTFGTN
Mvan_1899|M.vanbaalenii_PYR-1 NIQADPNVRVRLNGRWHRGTAHLLPEDDARARMRTLPGYNNVGVRTFGTN
MMAR_3562|M.marinum_M NIVDNPNIRIRQGTRWRTGVATLRPDDDVVARGRKFGRLGATVVKALETT
MUL_2795|M.ulcerans_Agy99 NIVDNPNIRIRQGTRWPTGVAALRPDDDVVARGRKFGRLGATVVKALETT
MAB_4636|M.abscessus_ATCC_1997 NIAANPNVRVRQGNRWRTGTAQFRPDDDVVARGRGFGVIGARVVKALETT
** : :*:* ** * * : *:** * : . *: : :
Mb1902c|M.bovis_AF2122/97 LLTIRVDLD-
Rv1871c|M.tuberculosis_H37Rv LLTIRVDLD-
MAV_2845|M.avium_104 LLTIRVDLD-
Mflv_4462|M.gilvum_PYR-GCK LLTIRVDLID
Mvan_1899|M.vanbaalenii_PYR-1 LLTVRVDLID
MMAR_3562|M.marinum_M PVSVRIDFTD
MUL_2795|M.ulcerans_Agy99 PVSVRIDFTD
MAB_4636|M.abscessus_ATCC_1997 PISVRIDFTD
:::*:*: