For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFGIIRPCRHRLGRELTVAWRAQLCGLCLALRDDYGQAARIATNYDGLVVSVLVEAQSVTPAPRRTAGPC PLRGMRRADVATGECARLAAVVSLALAAARVRDHIEDRDGVVGAAPIRPTARRIAQRWVRQGTDAGNALG FDTNVLVAAMDRQADLEANAVAGSSLLLVTEPTETAVAAAFEHTAVLAQRPANQAPLREIGRLFGRIAHL LDAVADYHDDLARGKWNPLTATATSIAAVRPLCDDALLGIELALADVDFTDGRLARRLLTREVRHSVSRT FSQLGYPGYEATPHGRQEPINFGAQPMSAGPYPGLPGEPAPNPDQSPKRGCWNTCTEDRCCDCTCDCCEG CCDDDCCCDCGDCADCCDCGDCCDCGDCCDCS
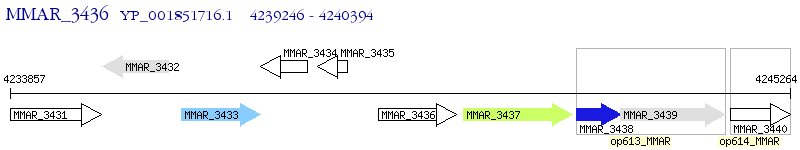
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3436 | - | - | 100% (382) | hypothetical protein MMAR_3436 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0396 | - | 7e-05 | 44.74% (38) | hypothetical protein Mflv_0396 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3436|M.marinum_M MFGIIRPCRHRLGRELTVAWRAQLCGLCLALRDDYGQAARIATNYDGLVV
Mflv_0396|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3436|M.marinum_M SVLVEAQSVTPAPRRTAGPCPLRGMRRADVATGECARLAAVVSLALAAAR
Mflv_0396|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3436|M.marinum_M VRDHIEDRDGVVGAAPIRPTARRIAQRWVRQGTDAGNALGFDTNVLVAAM
Mflv_0396|M.gilvum_PYR-GCK ----MDERDDDTSRAVVR--SLRTEDSYVVAG------------------
:::**. .. * :* : * : :* *
MMAR_3436|M.marinum_M DRQADLEANAVAGSSLLLVTEPTETAVAAAFEHTAVLAQRPANQAPLREI
Mflv_0396|M.gilvum_PYR-GCK ---------SVATQGSAVVAGSIATAGSAGVAGSIATARSAAVANSIATA
:** .. :*: . ** :*.. : . *: .* .:
MMAR_3436|M.marinum_M GRLFGRIAHLLDAVADYHDDLARGKWNPLTATATSIAAVRPLCDDALLGI
Mflv_0396|M.gilvum_PYR-GCK G-------------------------SAAVAQSIATAGSAAIANSILVVL
* .. .* : : *. .:.:. *: :
MMAR_3436|M.marinum_M ELALADVDFTDGRLARRLLTREVRHSVSRTFSQLGYPGYEATPHGRQEPI
Mflv_0396|M.gilvum_PYR-GCK SLACA-----------------------KCLACLG---------------
.** * : :: **
MMAR_3436|M.marinum_M NFGAQPMSAGPYPGLPGEPAPNPDQSPKRGCWNTCTEDRCCDCT-CDCCE
Mflv_0396|M.gilvum_PYR-GCK ---------------------------------------CIACTSCKACV
* ** *..*
MMAR_3436|M.marinum_M GCCDDDCCCDCGDCADCCDCGDCCDCGDCCDCS
Mflv_0396|M.gilvum_PYR-GCK GCIDCEDCIGCVGCIDCSGLRGAVGLRGVHAA-
** * : * .* .* **.. .. . . .