For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KSIAAARRVKVSADGHGVVSHAGMGLLRELADQTGLSEQVTGALSDTYKGPWVYAPGAVFADLAAAVADG ADCIHGVGQLCGDREHVFGAKASTTTMWRLVDERIDAVHLPKVRAARAAARATAWAGGAAPARGQPLCID IDATLVIDHSDNKADAAPTWKKSFGHHPLLAFLDRPEIAGGEALAGLLRKGNAGSNTAADHVTVLDQALD SLPADRRPDPDAEGAPAVVVRCDTAGATHKFAQACRDRGVGFSFGFPVDARVWDAVDTLNLAEGWYPAID AGGEIRDGAWVAEATSLVNLCTWPAGTRLVLRKERPHPGAQLRFTDIDGMRITAFITDTAPGVIPGQLAG LELRHRQHARVEDRIREAKAAGLANLPCHGFDPNAAWLEIVLAAADLVSWAQLIGFTDTPELARCEIASF RYRVLHAAARITRSARQLRLRIDATWRWATAIATAWRRIRAAFP*
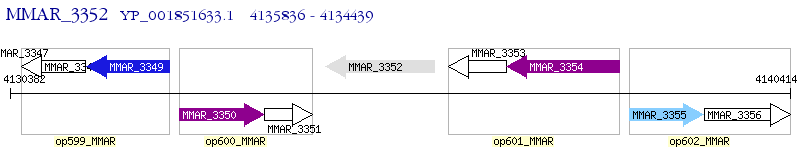
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3352 | - | - | 100% (465) | transposase for insertion sequence ISMyma02 |
| M. marinum M | MMAR_1468 | - | 0.0 | 100.00% (465) | transposase for insertion sequence ISMyma02 |
| M. marinum M | MMAR_1445 | - | 0.0 | 100.00% (465) | transposase for insertion sequence ISMyma02 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3048 | - | 0.0 | 82.44% (467) | transposase, IS4 family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_1972 | - | 0.0 | 79.87% (467) | putative transposase |
| M. smegmatis MC2 155 | MSMEG_6146 | - | 0.0 | 81.58% (467) | ISMsm3, transposase |
| M. thermoresistible (build 8) | TH_1997 | - | 1e-152 | 75.94% (345) | ISMsm3, transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5550 | - | 2e-15 | 23.73% (472) | transposase, IS4 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3048|M.gilvum_PYR-GCK ------------MKSIAAASRVK---VSADGHGVVSHAGMGLLRELADRT
MSMEG_6146|M.smegmatis_MC2_155 ------------MKSIAAASRVK---VSADGHGVVSHAGMGLLRELADRT
MAV_1972|M.avium_104 ------------MKNIAAGSRVK---VSADGHGVVSHAGMGMLRELADRT
MMAR_3352|M.marinum_M ------------MKSIAAARRVK---VSADGHGVVSHAGMGLLRELADQT
TH_1997|M.thermoresistible__bu ---------LNAVREVFPATREQRCWFHKQANVLAALPKSAHPSALAALK
Mvan_5550|M.vanbaalenii_PYR-1 MFSTHLSTQLRKALPVQVSHKFAASSAVFDDDHLVSCAGLVPVMTLAAQT
: . : : . :.: . ** .
Mflv_3048|M.gilvum_PYR-GCK GLSAQVTG--ALADTYRGPWTYAPGEVFADLAAAVADGADCIDGVGQLCG
MSMEG_6146|M.smegmatis_MC2_155 GLSAQVTG--ALADTYRGPWTYAPGEVFADLAAAVADGADCIDGVGQLCG
MAV_1972|M.avium_104 GLSAQVSA--ALADTYRGPWVYAPGDVFADLAAAVADGADCIDGVGQLCG
MMAR_3352|M.marinum_M GLSEQVTG--ALSDTYKGPWVYAPGAVFADLAAAVADGADCIHGVGQLCG
TH_1997|M.thermoresistible__bu EIYNAEDI--DKAQVAVKAFEVDFGAKYPKAVAKITDDLDTLLEFYRYPA
Mvan_5550|M.vanbaalenii_PYR-1 GLAQLLAAKVRITEPRIKSGSANPAPKLTTVVAGMCAGADCIDDLDLVRS
: :: . . . .* : . * : . .
Mflv_3048|M.gilvum_PYR-GCK DREHAFGAKASTTTMWRLVDERIDAAHLPAVRAARASAR------AAAWA
MSMEG_6146|M.smegmatis_MC2_155 DREHAFGAKASTTTMWRLVDERIDAAHLPAVRAARASAR------AAAWA
MAV_1972|M.avium_104 DREHVFGAKASTTTMWRLIDKRIDAARLPGVRAARAGAR------AAAWA
MMAR_3352|M.marinum_M DREHVFGAKASTTTMWRLVDERIDAVHLPKVRAARAAAR------ATAWA
TH_1997|M.thermoresistible__bu EHWIHLRTTNPIESTFATVRLRTKVTKGPGSRAAGLAMAYKLIDAAAXWD
Mvan_5550|M.vanbaalenii_PYR-1 GGMKTLFDGVYAPSTIGTLLREFTFGHARQLESVLREHLAG-----LCER
: : : . : .:.
Mflv_3048|M.gilvum_PYR-GCK AGAAPEPGGWLHIDIDATLVIDHSDNKHGATPTWKKSFGHHPLLAFLDRP
MSMEG_6146|M.smegmatis_MC2_155 AGAAPEPGGWLHIDIDATVVIDHSDNKHGATPTWKKTFGHHPLLAFLDRP
MAV_1972|M.avium_104 AGATPTGDGWLHIDIDATLVIDHSDNKTGATPTWKKTFGLHPLLAFLDRP
MMAR_3352|M.marinum_M GGAAPARGQPLCIDIDATLVIDHSDNKADAAPTWKKSFGHHPLLAFLDRP
TH_1997|M.thermoresistible__bu AAAAPADQDWLHIDLDATLVIDHSDNKAGATPTWKKTFGHHPLLAFLDRP
Mvan_5550|M.vanbaalenii_PYR-1 VQLLPGSDERAFVDIDSLLRPVYGRAKQGASYGHTKIAGKQILRKGLSPL
* :*:*: : :. * .*: .* * : * *.
Mflv_3048|M.gilvum_PYR-GCK EIAGGEALAGLLRSGNAGSNTASDHIIVLAQALAALPAPWRPDPSRGGGP
MSMEG_6146|M.smegmatis_MC2_155 EIAGGEALAGLLRSGNAGSNTASDHIIVLAQALAALPAPWRPDSSRGGDP
MAV_1972|M.avium_104 EISGGEALAGLLRTGNAGSNTASDHIIVLKQALASLPPRWRPHPGARADP
MMAR_3352|M.marinum_M EIAGGEALAGLLRKGNAGSNTAADHVTVLDQALDSLPADRRPDPDAEG--
TH_1997|M.thermoresistible__bu EIAGGEALGGLLRTSNAGANTASDHIIVLEQALAALPPAWQPDPDQPGDP
Mvan_5550|M.vanbaalenii_PYR-1 VTTISTETAAPVITGARLRAGKTNSGKGAARMIAQAVATARASG------
: . .. : .. :: : : . :.
Mflv_3048|M.gilvum_PYR-GCK DRPQVLVRCDTAGATHRFADACREQGVGFSFGYPVDARVQDAVDTLNLAQ
MSMEG_6146|M.smegmatis_MC2_155 DRPQVLVRCDTAGATHRFADACREQGVGFSFGYPVDARVQDAADTLNLAE
MAV_1972|M.avium_104 DASGVLVRCDTAGATHKFADACRAAGVGFSFGYPVDVRVQDAVDTLNLAD
MMAR_3352|M.marinum_M -APAVVVRCDTAGATHKFAQACRDRGVGFSFGFPVDARVWDAVDTLNLAE
TH_1997|M.thermoresistible__bu DKPKVLVRCDTAGATHKFAEACRTAGVGFSFGYPVDVRVQDAVDTLNLGS
Mvan_5550|M.vanbaalenii_PYR-1 VTGQILVRGDSAYGNSAVVAACRRAGARFSLVLTKTAAVAAAIESIS-ES
::** *:* .. .. *** *. **: . . * * :::. .
Mflv_3048|M.gilvum_PYR-GCK GWYPAIDTDGGLRD---GAWVAEATTLVN-LNSWPPGTRLILRKERPHPG
MSMEG_6146|M.smegmatis_MC2_155 GWYPAIDTDGGIRD---GAWVAEATTLVN-LNSWPPGTRLILRKERPHPG
MAV_1972|M.avium_104 GWYPAIDTDGGIRE---GAWVAEATDLVN-LSSWPAGTRLILRKERPHPG
MMAR_3352|M.marinum_M GWYPAIDAGGEIRD---GAWVAEATSLVN-LCTWPAGTRLVLRKERPHPG
TH_1997|M.thermoresistible__bu CWYPAIDTRGGIRD---GAWVAEATDLVN-LESWPPGTRLILRKERPHPG
Mvan_5550|M.vanbaalenii_PYR-1 AWVP-VKYPGAVRDPDTGAWISDAEVAETTYTAFSSTKTPVTARLIVRRV
* * :. * :*: ***:::* . ::.. . : : :
Mflv_3048|M.gilvum_PYR-GCK AQLRFTDTDG--MRVTAFITDTPTGVVAGQVAGLELRHRQHARVEDRIRE
MSMEG_6146|M.smegmatis_MC2_155 AQLRFTDTDG--MRVTAFITDTPTGVVAGQVAGLELRHRQHARVEDRIRE
MAV_1972|M.avium_104 AQLRFTDADG--MRVTAFITDTEPGVIAGQVAGLELRHRQHARVEDRIRE
MMAR_3352|M.marinum_M AQLRFTDIDG--MRITAFITDTAPGVIPGQLAGLELRHRQHARVEDRIRE
TH_1997|M.thermoresistible__bu AQLRFTDADG--MRVTAFITDTPHGVVPGQVAGLELRHRQHARVEDRIRE
Mvan_5550|M.vanbaalenii_PYR-1 KDARFLDALFPVWRYHPFFTDSDE-----PVDASDITHRRHAIIETVFAD
: ** * * .*:**: : . :: **:** :* : :
Mflv_3048|M.gilvum_PYR-GCK LKNTGLRNLPCHGFWANAAWLEIVLAAADLVTWARLIGFTSDPTLARAEI
MSMEG_6146|M.smegmatis_MC2_155 LKNTGLRNLPCHGFWANAAWLEIVLAATDLVTWARLIGFTSDPTLARAEI
MAV_1972|M.avium_104 LKATGLRNLPCHSFWANAAWLEIVLAAADLVTWARLIGFRAHPGLARAEI
MMAR_3352|M.marinum_M AKAAGLANLPCHGFDPNAAWLEIVLAAADLVSWAQLIGFTDTPELARCEI
TH_1997|M.thermoresistible__bu LKATGLRNLPCHSFWANAAWLEIVLAAADLVTWTRLIGFRNQPGLARAEI
Mvan_5550|M.vanbaalenii_PYR-1 LIDGPLAHMPSGRFGANSAWILCAAIAYNLL---RAVGVLAGGVHAVARG
* ::*. * .*:**: . * :*: : :*. * ..
Mflv_3048|M.gilvum_PYR-GCK ATFRYRVLHVAARITRGARQLRLRIDATWRWATSIATAWQTIRTAFG---
MSMEG_6146|M.smegmatis_MC2_155 ATFRYRVLHVAARITRGARQLRLRIDATWRWASAIATAWQHLRTAFG---
MAV_1972|M.avium_104 ATFRYRVLHVAARITRGARQQRLRIDATWRWASAIATAWQHLRTAFG---
MMAR_3352|M.marinum_M ASFRYRVLHAAARITRSARQLRLRIDATWRWATAIATAWRRIRAAFP---
TH_1997|M.thermoresistible__bu NTFRYRVLHVAARITRGARQLRLRIDATWRWAAAIVTAWQHLRTAFG---
Mvan_5550|M.vanbaalenii_PYR-1 ATLRRKFVNIPARLARPQRRAILHLPTHWPWAQQWLTLWRNTIGHSPPTA
::* :.:: .**::* *: *:: : * ** * *:
Mflv_3048|M.gilvum_PYR-GCK ---
MSMEG_6146|M.smegmatis_MC2_155 ---
MAV_1972|M.avium_104 ---
MMAR_3352|M.marinum_M ---
TH_1997|M.thermoresistible__bu ---
Mvan_5550|M.vanbaalenii_PYR-1 ATH