For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIFDLDGTLTDSAHGIVASFRHALNHIGAAVPDGDLVAQIVGPPMDDTFHAMELGDRVEDAIAAFRAEYG ARGWAMNTLFDGIAPLLADLRAAGVRLAVATCKLEPTARRVLAHFGLDAHFEVIAGATGDGARRSKTEVL AHALAQLHPLPERVLMVGDRSHDVEGAAAHGIDTVVVGWGYGKADFAAGTHSGAALHAGTVDELREALGV
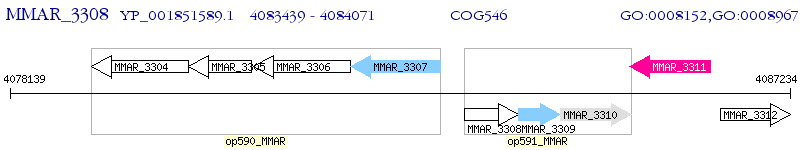
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3308 | - | - | 100% (210) | hypothetical protein MMAR_3308 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2257 | - | 4e-90 | 75.24% (210) | hypothetical protein Mb2257 |
| M. gilvum PYR-GCK | Mflv_2889 | - | 2e-83 | 71.43% (210) | HAD family hydrolase |
| M. tuberculosis H37Rv | Rv2232 | - | 4e-90 | 75.24% (210) | hypothetical protein Rv2232 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1901c | - | 5e-65 | 64.17% (187) | hypothetical protein MAB_1901c |
| M. avium 104 | MAV_2207 | - | 1e-91 | 80.48% (210) | hypothetical protein MAV_2207 |
| M. smegmatis MC2 155 | MSMEG_4308 | - | 4e-80 | 69.52% (210) | 5'-nucleotidase |
| M. thermoresistible (build 8) | TH_1464 | - | 2e-78 | 69.52% (210) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1320 | - | 2e-73 | 99.26% (135) | hypothetical protein MUL_1320 |
| M. vanbaalenii PYR-1 | Mvan_3621 | - | 1e-78 | 68.10% (210) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2889|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3621|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3308|M.marinum_M --------------------------------------------------
MUL_1320|M.ulcerans_Agy99 --------------------------------------------------
MAV_2207|M.avium_104 --------------------------------------------------
Mb2257|M.bovis_AF2122/97 MSSPRERRPASQAPRLSRRPPAHQTSRSSPDTTAPTGSGLSNRFVNDNGI
Rv2232|M.tuberculosis_H37Rv MSSPRERRPASQAPRLSRRPPAHQTSRSSPDTTAPTGSGLSNRFVNDNGI
TH_1464|M.thermoresistible__bu --------------------------------------------------
MSMEG_4308|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1901c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2889|M.gilvum_PYR-GCK MTDMLDGTRRVAP------------VGRGDLVLFDLDGTLTDSAEGIVAS
Mvan_3621|M.vanbaalenii_PYR-1 MTDMLDGTRDLAP------------VERGDVVIFDLDGTLTDSAEGIVAS
MMAR_3308|M.marinum_M -------------------------------MIFDLDGTLTDSAHGIVAS
MUL_1320|M.ulcerans_Agy99 --------------------------------------------------
MAV_2207|M.avium_104 MTGTTSADPAAP---------------AAQLVIFDLDGTLTDSAEGIVAS
Mb2257|M.bovis_AF2122/97 VTDTTASGTNCPPPPRAAARRASSPGESPQLVIFDLDGTLTDSARGIVSS
Rv2232|M.tuberculosis_H37Rv VTDTTASGTNCPPPPRAAARRASSPGESPQLVIFDLDGTLTDSARGIVSS
TH_1464|M.thermoresistible__bu VTETRDDTAAVPAGAVDAARRPVDPALRPELVIFDLDGTLTDSAEGIVSS
MSMEG_4308|M.smegmatis_MC2_155 MTDVVTGRVSELTP-----------DTRPQLVLFDLDGTLTDSAEGIVSS
MAB_1901c|M.abscessus_ATCC_199 -----------------------------MLVIFDLDGTLTDSAPGIVSS
Mflv_2889|M.gilvum_PYR-GCK FRHALQSVGAAVPDGDLVSKIVGPPMPLTFQRMGLGDLVDDAIAAYRADY
Mvan_3621|M.vanbaalenii_PYR-1 FRHALHAVGAPVPDGDLVSRIVGPPMHVTLQQMGLGEHADAAIAAYRADY
MMAR_3308|M.marinum_M FRHALNHIGAAVPDGDLVAQIVGPPMDDTFHAMELGDRVEDAIAAFRAEY
MUL_1320|M.ulcerans_Agy99 --------------------------------------------------
MAV_2207|M.avium_104 FLHALESIGAPVPPGDLVAQIVGPPMDDTFRSMQLGEDAEAAIAAFRAEY
Mb2257|M.bovis_AF2122/97 FRHALNHIGAPVPEGDLATHIVGPPMHETLRAMGLGESAEEAIVAYRADY
Rv2232|M.tuberculosis_H37Rv FRHALNHIGAPVPEGDLATHIVGPPMHETLRAMGLGESAEEAIVAYRADY
TH_1464|M.thermoresistible__bu FRHALASVGAAVPDGDLAARIIGPPMHHTLRSLGLGDQTDAAIDAYRQDY
MSMEG_4308|M.smegmatis_MC2_155 FRHALQAVGAPVPDGDLASRIVGPPMHHTLTSLGLGDQADEAIAAYRADY
MAB_1901c|M.abscessus_ATCC_199 FRHALSHVGAPEPTGDIASRVVGPPMHITLGSMGLGERADDAIAAYRADY
Mflv_2889|M.gilvum_PYR-GCK TTRGWSMNRPFDGIPALLADLQAAGVRMAVATSKAETTAQRILAHFGLDG
Mvan_3621|M.vanbaalenii_PYR-1 VSRGWAMNQPFTGIPALLADLQAAGVRMAVATSKAEPTAQRILAHFGLDG
MMAR_3308|M.marinum_M GARGWAMNTLFDGIAPLLADLRAAGVRLAVATCKLEPTARRVLAHFGLDA
MUL_1320|M.ulcerans_Agy99 ------MNTLFDGIAPLLADLRAAGVRLAVATCKLEPTARRVLAHFGLDA
MAV_2207|M.avium_104 GARGWAMNTLFDGIEALLADLRAAGVRLAVATSKLEPTARRILAHFGLDH
Mb2257|M.bovis_AF2122/97 SARGWAMNSLFDGIGPLLADLRTAGVRLAVATSKAEPTARRILRHFGIEQ
Rv2232|M.tuberculosis_H37Rv SARGWAMNSLFDGIGPLLADLRTAGVRLAVATSKAEPTARRILRHFGIEQ
TH_1464|M.thermoresistible__bu RARGWAMNRVYDGIPELLADLRAAEVRLAVATSKAEPTAQRILAHFGLAD
MSMEG_4308|M.smegmatis_MC2_155 TSRGWAMNSLFDGIPALLSDLRAAGVRLAVATSKAEPTARRILEHFELDG
MAB_1901c|M.abscessus_ATCC_199 GERGWAVNEMFDGIRDVLTRLREAGVPMVVATSKSEPIAARILEHFGLAE
:* : ** :*: *: * * :.***.* *. * *:* ** :
Mflv_2889|M.gilvum_PYR-GCK HFEVIAGASNDGTRAAKSDVVAHALAQLAPVPDN-VVMIGDRSHDVEGAA
Mvan_3621|M.vanbaalenii_PYR-1 HFEVIAGAGADGTRAAKADVVARALEQLAPVPQR-MVMIGDRSHDVEGAA
MMAR_3308|M.marinum_M HFEVIAGATGDGARRSKTEVLAHALAQLHPLPER-VLMVGDRSHDVEGAA
MUL_1320|M.ulcerans_Agy99 HFEVIAGATGDGARRSKTEVLAHALAQLHPLPER-VLMVGDRSHDVEGAT
MAV_2207|M.avium_104 HFEVIAGASPDGSRKAKVEVLAHALAQLEPLPER-VLMVGDRSHDVHGAA
Mb2257|M.bovis_AF2122/97 HFEVIAGASTDGSRGSKVDVLAHALAQLRPLPER-LVMVGDRSHDVDGAA
Rv2232|M.tuberculosis_H37Rv HFEVIAGASTDGSRGSKVDVLAHALAQLRPLPER-LVMVGDRSHDVDGAA
TH_1464|M.thermoresistible__bu HFEFIAGASVDGSRAAKADVLARALQALQPLPER-VLMVGDRCHDVEGAA
MSMEG_4308|M.smegmatis_MC2_155 YFEVIAGASPDGTRSLKSEVVAHALRQLEPLPDR-VLMVGDRMHDVEGAA
MAB_1901c|M.abscessus_ATCC_199 FFQVIAGASPDGVRSSKADVIKHALSQLDDVPPTGVVMVGDRVHDVEGAA
.*:.**** ** * * :*: :** * :* ::*:*** ***.**:
Mflv_2889|M.gilvum_PYR-GCK VHGIATIVVGWGYGRTDFAG----RESGALAHVTDVDDLREVLGV
Mvan_3621|M.vanbaalenii_PYR-1 VHGIGTIVVGWGYGGRDFAD----GDGGALAHVADIDDLREVLGV
MMAR_3308|M.marinum_M AHGIDTVVVGWGYGKADFAAG---THSGAALHAGTVDELREALGV
MUL_1320|M.ulcerans_Agy99 AHGIDTVVVGWGYGKADFAAG---THSGAALHAGTVDELREALGV
MAV_2207|M.avium_104 AHGIDTVVVGWGYGHAD-------SHPDGVTRAATIDELRRALGV
Mb2257|M.bovis_AF2122/97 AHGIDTVVVGWGYGRADFIDK---TSTTVVTHAATIDELREALGV
Rv2232|M.tuberculosis_H37Rv AHGIDTVVVGWGYGRADFIDK---TSTTVVTHAATIDELREALGV
TH_1464|M.thermoresistible__bu AHGIDTVVVGWGYGRDDFAHE---DAIPAYAHVATVAELREVLSV
MSMEG_4308|M.smegmatis_MC2_155 EHGIDTVVVGWGYGGADFSGP---AAVVAHAHVETVEALRGVLGV
MAB_1901c|M.abscessus_ATCC_199 AHGIGTVVVDWGYGATDFEGDPGQLAEWIIGRVSTPEQLLEELDV
*** *:**.**** * :. * *.*