For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSIGIHDLSIATAHYVLDHATLAEHHGVDVNKYLIGLGQQQMSIVAPDEDIVTLAAAAADPIIKRHGSQK IRTIVIGTETGVDQSKSAGIWVSSLLGLPSSARVLEVKQACYGATGALQLALALVHRDPTQQVLVIAADV ARYDLDSPGEPTQGAAAAAMLVSADPALLRLEEPTGIYTADIMDFWRPNYRSTALVDGKASVTAYMEAAS GAWKDYTERGGRAFGEFAAFCYHQPFTKMAYKAHKQLAAEAGEDASGAAVQAAVGNTVEYNRRIGNSYTA SLYLALAALLDQADDLSDQPIAMLSYGSGCVAELFAGTVTPGYQQHLRTDQHRAALETRIPLSYEHYRRL HNLTLPTNGNHHSLPVETSRPFRLTAISEHKRMYGAV*
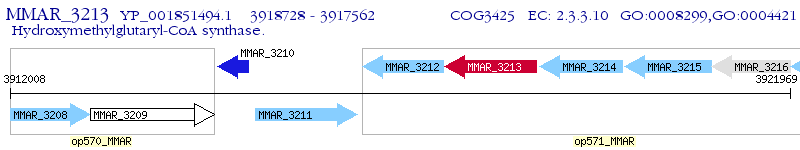
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3213 | pksG | - | 100% (388) | hydroxymethylglutaryl-coenzyme A synthase, PksG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_5080 | - | 4e-06 | 24.15% (236) | hypothetical protein MAV_5080 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3213|M.marinum_M MVSIGIHDLSIATAHYVLDHATLAEHHG------VDVNKYLIGLGQQQMS
MAV_5080|M.avium_104 -MSSGLLSYASYLPRYRLSGQDIGVRHGNRVVASYDEDSTTMAVAAASSA
:* *: . : .:* *. :. :** * :. :.:. . :
MMAR_3213|M.marinum_M IVAPDEDIVTLAAAAADPIIKRHGSQKIRTIVIGTETGVDQSKSAGIWVS
MAV_5080|M.avium_104 LMHSAVSPRALYFATSSPAYADKTNATAIHAALGLPADMPAADMCGTARS
:: . . :* *::.* : . . .:* :.: :. .* *
MMAR_3213|M.marinum_M SLLGLPSSARVLEVKQACYGATGALQLALALVHRDPTQQVLVIAADVARY
MAV_5080|M.avium_104 AFAALTLAASSGGLVVAADVRVGRPGSADEKLGGDGAAALIFGEGDVIAE
:: .*. :* : *. .* * : * : ::. .**
MMAR_3213|M.marinum_M DLDSPGEPTQGAAAAAMLVSADPALLRLEEPTGIYTADIMDFWRPNYRST
MAV_5080|M.avium_104 YLASRSLTAEFLDRWRDPTSRTGQQWEERFGAQHYTALIRDAAREALDAA
* * . .:: .* . . : *** * * * ::
MMAR_3213|M.marinum_M ALVDGKASVTAYMEAASGAWKDYTERGGR------AFGEFAAFCYHQPFT
MAV_5080|M.avium_104 GIADPDHVVVACPNSGVVKRAATLVKGQKTVATSVGFGGSADALIALCGT
.:.* . *.* ::. :* : .** * *
MMAR_3213|M.marinum_M KMAYKAHKQLAAEAGEDASGAAVQAAVG--------NTVEYNRRIGNSYT
MAV_5080|M.avium_104 LDVAEAGDTILVLSATDGCDAMVLRATSRLPRARQPRPLNDQRAEGTPVE
. :* . : . :. *...* * *.. ..:: :* *..
MMAR_3213|M.marinum_M ASLYLALAALLDQADDLSDQP----------IAMLSYGSG-------CVA
MAV_5080|M.avium_104 YVTYLAWRGLVTMEPPRRPEPERPAAPPASRARLWKFGMTGARCRACGFM
*** .*: :* : .:* .
MMAR_3213|M.marinum_M ELFAGTVTPGYQQHLRTDQHRAALETRIPLSYEHYRRLHNLTLPTN----
MAV_5080|M.avium_104 HLPPVRVCRSCNAIDEMDPQSAAGLRGTVVTYTVDRLAYSPSPPMVQAVV
.* . * . : . * : ** ::* * :. : *
MMAR_3213|M.marinum_M ----GNHHSLPVETSR------------PFR--LTAISEHKRMYGAV---
MAV_5080|M.avium_104 DVDGGGRCTLEVADARVEDLRVGTRVRFSFRRLFTASDVHNYFWKVVQDD
*.: :* * :* .** :** . *: :: .*
MMAR_3213|M.marinum_M --
MAV_5080|M.avium_104 AE