For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEPCGADARATATQLSPTQDNYPDRLDWPTIRIAGVCLIAAIMANLDISIVTVAQRTFTVAFHSTQATVA WTVAGYMLGMATATPMTGWAADRLGAKRLFMGAVATFTLGSMLCASAPNIGLLITFRVVQGIGGGVLGPL VLAIVTHQAGPRRLGRLLAVGAIPMLTAPMLGPILGGWLIDSYGWQWIFLINVPAGLLAFGLAAILVPED PPKPSERFDFIGMLLLLPGIAMLLLGVSAIPGSGTVTDHRVWVPAISGAVLITAFALHAWYRTDHPLIDL RLFTDRVVRLANLALLLYVAGAAGASLLLPSYFQQLLHQTPMRSGLMMVPIGFGAMLTMPLTGAFMDSRG PRKVVLIGLTLIAAGTGTFVFGVANEADYLPTLLAGLTIAGMGLGCTGLLLAASVMRVLAPHQIARGSAL ISVNQQISGSIGAALMSMILTNQFCRSHTISTANNMAVLRENAERHVVPIAPSAVPARAVGPDFLVELQH DLSHAYTLTFAVAAVLGALAYFPAAFLPAKPTTIAPGEVDVPG
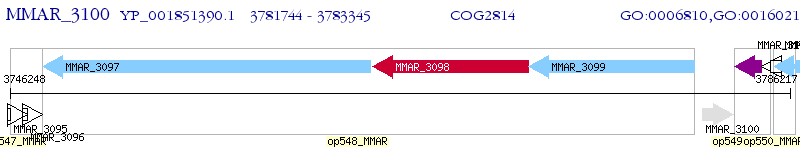
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3100 | ermB_1 | - | 100% (533) | integral membrane drug efflux protein, ErmB_1 |
| M. marinum M | MMAR_4905 | - | e-170 | 59.49% (506) | integral membrane drug efflux protein |
| M. marinum M | MMAR_2515 | - | e-168 | 57.51% (506) | integral membrane drug efflux protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 1e-168 | 57.20% (500) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_2221 | - | 6e-52 | 29.23% (479) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 1e-168 | 57.20% (500) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_01562 | - | 4e-29 | 25.95% (420) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_1396 | - | 6e-49 | 30.30% (406) | EmrB/QacA family drug resistance transporter |
| M. avium 104 | MAV_0729 | - | 1e-170 | 57.80% (519) | EmrB efflux protein |
| M. smegmatis MC2 155 | MSMEG_5046 | - | 7e-47 | 31.42% (401) | drug transporter |
| M. thermoresistible (build 8) | TH_3965 | - | 6e-47 | 28.88% (412) | PUTATIVE drug transporter |
| M. ulcerans Agy99 | MUL_0501 | - | 1e-168 | 58.70% (506) | integral membrane drug efflux protein |
| M. vanbaalenii PYR-1 | Mvan_4474 | - | 4e-53 | 30.77% (416) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0805c|M.bovis_AF2122/97 --------------MLGNAMVEACPAEGDAPVPITPAGRPRSGQRSYPDR
Rv0783c|M.tuberculosis_H37Rv --------------MLGNAMVEACPAEGDAPVPITPAGRPRSGQRSYPDR
MUL_0501|M.ulcerans_Agy99 --------------MLSTAMDEAHCTPAEVAPPDTRTGTSSPGEHVYPDR
MAV_0729|M.avium_104 --------------MLSGAMEKTR--SAAFSVPLATASGPSLRDDEYPDK
MMAR_3100|M.marinum_M -------------------MEPCG------ADARATATQLSPTQDNYPDR
Mflv_2221|M.gilvum_PYR-GCK --------------------------MFDTVTSRAAGT------------
Mvan_4474|M.vanbaalenii_PYR-1 --------------------------MFETVTARVRG-------------
MSMEG_5046|M.smegmatis_MC2_155 --------------------------MPSSLNARIAGLLPSRVGALRPQD
TH_3965|M.thermoresistible__bu ----------------VTSVDAGRSTMSEVFTAYAPRS------------
MAB_1396|M.abscessus_ATCC_1997 --------------------------MRPEYPR-----------------
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mb0805c|M.bovis_AF2122/97 LDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTG
Rv0783c|M.tuberculosis_H37Rv LDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTG
MUL_0501|M.ulcerans_Agy99 LDAGLLRIAGVCVLASLMSVLDATVVGIAQRTFIIEFDSTQAVVAWTMTG
MAV_0729|M.avium_104 LDAALFRIAGVCGLACIMAVLDSTVVAVAQRTFIAQFGANQAIVSWTIAG
MMAR_3100|M.marinum_M LDWPTIRIAGVCLIAAIMANLDISIVTVAQRTFTVAFHSTQATVAWTVAG
Mflv_2221|M.gilvum_PYR-GCK --TNPWPALWALLIGFFMILVDATIVAVANPAIMASLNAGYDAVIWVTSA
Mvan_4474|M.vanbaalenii_PYR-1 --ADPWPALWALLIGFFMILVDATIVPVANPAIMAQLGADYDAVIWVTSA
MSMEG_5046|M.smegmatis_MC2_155 GTGNPWNALWAMMFGFFMILVDATIVSVANPTIMVELGADYDGVIWVTSA
TH_3965|M.thermoresistible__bu AAANPWHALWAMLIGFFMILVDATIVPVANLEIMADLGADYTAVIWVTSA
MAB_1396|M.abscessus_ATCC_1997 ----PWITLWVVLFGLFMILLDSTIVSVANPAIKAGFGADYSSTVWVTSA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
. .: :* ::. :* : : * :.
Mb0805c|M.bovis_AF2122/97 YMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVAPNILLLII
Rv0783c|M.tuberculosis_H37Rv YMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVAPNILLLII
MUL_0501|M.ulcerans_Agy99 YTLALATVIPVAGWAADRFGTKRLWMGSVLAFALGSLLCALAPNILSLIF
MAV_0729|M.avium_104 YMLAFATVIPITGWAADRFGTKRLFMGSVLIFTLGSLLCAVAPNILLLIL
MMAR_3100|M.marinum_M YMLGMATATPMTGWAADRLGAKRLFMGAVATFTLGSMLCASAPNIGLLIT
Mflv_2221|M.gilvum_PYR-GCK YLLAYAVPLLVSGRLGDRYGPKNVYMVGLTVFTVASLWCGLSDSIGMLIA
Mvan_4474|M.vanbaalenii_PYR-1 YLLAYAVPLLVSGRLGDRYGPKTVYLVGLAVFTAASLWCGLSDSIGMLVA
MSMEG_5046|M.smegmatis_MC2_155 YLLAYAVPLLVTGRLGDMYGPKNLYLLGLGVFTAASLWCGLAGSIEMLIA
TH_3965|M.thermoresistible__bu YLLAYAVPLLIAGRLGDRFGPRTMYLIGLAIFTAASLWCGLAGTIGMLIA
MAB_1396|M.abscessus_ATCC_1997 YLLGYAVPLLITGRLGDQFGPRSMYLSGLAVFTAASLWCGLAGSIGWLIA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
* * . * .* * : :: .: *: .*: *. : *:
Mb0805c|M.bovis_AF2122/97 FRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPMLLGPVGGPI
Rv0783c|M.tuberculosis_H37Rv FRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPMLLGPVGGPI
MUL_0501|M.ulcerans_Agy99 FRVLQGVGGGMLMPLGFIILTRAAGP-KRLGRLMAAIGIPMLLGPIGGPI
MAV_0729|M.avium_104 FRVVQGVGGGMLLPLSFVILTREAGP-KRVGRLMAVGGIPILLGPIGGPI
MMAR_3100|M.marinum_M FRVVQGIGGGVLGPLVLAIVTHQAGP-RRLGRLLAVGAIPMLTAPMLGPI
Mflv_2221|M.gilvum_PYR-GCK ARVVQGVGAALLTPQTMTVITRTFPA-HRRGVAMSVWGATAGVATLVGPL
Mvan_4474|M.vanbaalenii_PYR-1 ARVLQGVGAALLTPQTLTVITRTFPA-HNRGVAMSVWGATAGVATLAGPL
MSMEG_5046|M.smegmatis_MC2_155 ARVVQGIGAALLTPQTLSAITRIFPA-NRRGVAMSVWGATAGVATLVGPL
TH_3965|M.thermoresistible__bu ARVLQGLGAALLTPQTLSMITRIFPA-DRRGVPMGVWGATAGVATLIGPL
MAB_1396|M.abscessus_ATCC_1997 ARVVQGIGAAMLTPQTLTVVQRVFPP-ERRGTAMGVWGAVAGIATLVGPV
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
*::**.*..: * : : . .. . ..: * :
Mb0805c|M.bovis_AF2122/97 LGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASENFDYMGLL
Rv0783c|M.tuberculosis_H37Rv LGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASENFDYMGLL
MUL_0501|M.ulcerans_Agy99 LGGWLISSFGWHWIFLVNLPIGLAAFALAAITFPADRPVPSESFDVIGVL
MAV_0729|M.avium_104 LGGWLIGAYGWKWIFLINLPIGLTAFALAALLFPKDRSAPSEALDITGAL
MMAR_3100|M.marinum_M LGGWLIDSYGWQWIFLINVPAGLLAFGLAAILVPEDPPKPSERFDFIGML
Mflv_2221|M.gilvum_PYR-GCK AGGVLVDSLGWQWIFIVNVPVGIVGLVMALRLVPALPNPVRQGFDVPGVV
Mvan_4474|M.vanbaalenii_PYR-1 AGGVLVDSLGWQWIFIVNLPVGVLGFALAVWLVPSLPTARAQRFDLPGVV
MSMEG_5046|M.smegmatis_MC2_155 AGGVLVGGLGWQWIFFVNVPVGIVGIALAVWLVPTLPTTRHH-FDIPGIV
TH_3965|M.thermoresistible__bu AGGVLVDGLGWQWIFIVNVPIGIAGLVLGARLMPRLP-RQDQGFDVLGMV
MAB_1396|M.abscessus_ATCC_1997 AGGLLVDGWGWQWIFYVNVPVGVLGIILGAIYIPRMPTHRHR-LDLLGMA
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK-LDAAGAI
** * *: * :*:* *: : :. . . :* *
Mb0805c|M.bovis_AF2122/97 LLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFVAHSWYRTE
Rv0783c|M.tuberculosis_H37Rv LLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFVAHSWYRTE
MUL_0501|M.ulcerans_Agy99 LLSPGLAAFLLALSLIPDRGTVADRYVLIPVIAGLSLIGGFAWHAWHRAD
MAV_0729|M.avium_104 LLSPGVAIFLCGVCSIPGRHTVADRYVLVPALVGLVLIAAFILHAWYRTE
MMAR_3100|M.marinum_M LLLPGIAMLLLGVSAIPGSGTVTDHRVWVPAISGAVLITAFALHAWYRTD
Mflv_2221|M.gilvum_PYR-GCK LSGVGLFLIVFALQE--GQSHGWALWIWATVLAGVAVMAAFVYWQSINTA
Mvan_4474|M.vanbaalenii_PYR-1 LSGVALFLIVFALQE--GQSHDWAPWIWATIGIGIAVMAAFLYWQSVNTG
MSMEG_5046|M.smegmatis_MC2_155 LSAVGMFLIVFALQE--GQSKDWAPWVWGTATLGVGVMAAFFFWQSVNTR
TH_3965|M.thermoresistible__bu LSAVGLFLIVFALQE--AQTHRWAPWIWGTLAGGAGVMAMFVIWQSANRR
MAB_1396|M.abscessus_ATCC_1997 LSAVGMFAFVFALQE--GESFQWAPGVWAMMAAGLVVLGVFVWWQSANRG
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAEN-
* . : .. : : .
Mb0805c|M.bovis_AF2122/97 HPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQSPMQS
Rv0783c|M.tuberculosis_H37Rv HPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQSPMQS
MUL_0501|M.ulcerans_Agy99 HPLIDLHLFNNSVVTQANLTLLAFAAAYFGSSLLIPTYLQQVLHQTPMQS
MAV_0729|M.avium_104 HPLIDLRLFRNPVVTQVNVTLLVFAAASVGVGLLVPSYFQIVGHETPMQS
MMAR_3100|M.marinum_M HPLIDLRLFTDRVVRLANLALLLYVAGAAGASLLLPSYFQQLLHQTPMRS
Mflv_2221|M.gilvum_PYR-GCK GALIPLVIFRDRNFSMSNLGVATIGFVATGMILPLMFFAQAVCGLTPTRA
Mvan_4474|M.vanbaalenii_PYR-1 EPLIPLIIFGDRNFSMSNLGVATIGFVATGMILPLMFYAQSVCGLTPTQA
MSMEG_5046|M.smegmatis_MC2_155 EPLIPLRVFADRDFTLANIGVAVIGFAVTAMILPLMFYLQTVCGLSPIRS
TH_3965|M.thermoresistible__bu HPLVPLQIFADRNFMLANVGVATIGFVVTAMVLPAMFFAQAVCGLSPTGS
MAB_1396|M.abscessus_ATCC_1997 EPLIPLRLFADRNFSLAGVGIASMSFCVISTGLPMMFYTQLALGFSPTKA
MLBr_01562|M.leprae_Br4923 -PVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRA
.:: : :* : : : . : : * :. :
Mb0805c|M.bovis_AF2122/97 GVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGLGTFAFGVA
Rv0783c|M.tuberculosis_H37Rv GVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGLGTFAFGVA
MUL_0501|M.ulcerans_Agy99 GLYLIPQGLGAMLTMPIASAFMDRRGPGKSVLLGIVLIAVGLAMFTFGVA
MAV_0729|M.avium_104 GLHMLPIGVGAVLTMPLGGAVMDKHGPGKIVLTGLPLMAVGLAVFTYGVA
MMAR_3100|M.marinum_M GLMMVPIGFGAMLTMPLTGAFMDSRGPRKVVLIGLTLIAAGTGTFVFGVA
Mflv_2221|M.gilvum_PYR-GCK ALLTAPMAVATGVLAPFVGKLVDRAHPRSVVGFGFSLMAIGVTWLSIEMT
Mvan_4474|M.vanbaalenii_PYR-1 ALLTAPMAVATGVLAPLVGKLVDRSHPRPVVGFGFALMAIGLTWLSIEMT
MSMEG_5046|M.smegmatis_MC2_155 ALVTAPTAIASGVLAPFVGRIVDRVHPMPVIGFGFSAMAIGLTWLSIEMA
TH_3965|M.thermoresistible__bu ALLIAPMAVASGVLAPMAGRIVDRSHPTPVVGFGFSVVAVGLTWLAVEMA
MAB_1396|M.abscessus_ATCC_1997 ALTQAPTAIVSGILAPLAGWLVDRVRPSILIGGGIALMIASTLWWTLLMR
MLBr_01562|M.leprae_Br4923 GIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMN
.: * . : . . .:. * * : . :
Mb0805c|M.bovis_AF2122/97 RQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIARGSTLISVN
Rv0783c|M.tuberculosis_H37Rv RQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIARGSTLISVN
MUL_0501|M.ulcerans_Agy99 TRAQYLPTLLVGLAIMGMGTGCTMMPLAGAAVLTLKPREIARGSTLISVT
MAV_0729|M.avium_104 RQAAYSPVLVCGLAIMGLGIGLTTTPLSAALMQALAPHQVARGTTLISVN
MMAR_3100|M.marinum_M NEADYLPTLLAGLTIAGMGLGCTGLLLAASVMRVLAPHQIARGSALISVN
Mflv_2221|M.gilvum_PYR-GCK PTTPIWRLLLP-LTAMGAAMAFIWSPLAATATRNLPPHLAGAGSGVYNTT
Mvan_4474|M.vanbaalenii_PYR-1 PSTPIWRLVVP-LTAMGVAMAFIWSPLAATATRNLPPQLAGAGSGVYNTT
MSMEG_5046|M.smegmatis_MC2_155 PTTPIWRLLLP-QLLMGIGMAFIWSPLAATATRHLPPDLAGAGSGVYNTT
TH_3965|M.thermoresistible__bu PTTPIWRLVLP-FVAVGVGMAFIWSPLATTATRQLPLHLSGAGSGVYNTT
MAB_1396|M.abscessus_ATCC_1997 PETQMWQLMLP-AAGIGAAMACIWGPLATTATRNLPPSVAGAGSGVYNTL
MLBr_01562|M.leprae_Br4923 RGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALML
. : * . . *: : : . : :
Mb0805c|M.bovis_AF2122/97 QQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAAVDPSS
Rv0783c|M.tuberculosis_H37Rv QQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAAVDPSS
MUL_0501|M.ulcerans_Agy99 QQVGGSIGTALMATILTNQFNRSDNIWVANKVATLEQNAAARGVTVDPSG
MAV_0729|M.avium_104 QQVGGSIGAALMAVILTNQFNRNPALMAANEAAGMHPVTGKRGLPVDPST
MMAR_3100|M.marinum_M QQISGSIGAALMSMILTNQFCRSHTISTANNMAVLRENAERHVVPIAPSA
Mflv_2221|M.gilvum_PYR-GCK RQVGSVLGSAGMAALMSSELS--------TKIPG---ADAVPQSEGEVAA
Mvan_4474|M.vanbaalenii_PYR-1 RQVGSVLGSAAMAALLASQLS--------AKIPG---EAPVP-VEGQAGQ
MSMEG_5046|M.smegmatis_MC2_155 RQVGSVLGSAGIAAFMTWRIT--------DEVPGMQDAAPAGESGGAVEQ
TH_3965|M.thermoresistible__bu RQIGSVLGSAGMAAFLTGRLS--------AELPFG--ATAVP-AGEQTRV
MAB_1396|M.abscessus_ATCC_1997 RQVGGVVGSSAMGALMTARLT--------ATMPA----MPPGSGSTPEAA
MLBr_01562|M.leprae_Br4923 QSLGGPLVLAVIQAVITSRTLY------------------LGGTTGPVKF
:.:.. : : : .:: .
Mb0805c|M.bovis_AF2122/97 LPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQ----
Rv0783c|M.tuberculosis_H37Rv LPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQ----
MUL_0501|M.ulcerans_Agy99 LPPQALTPDFADNVMHDLSHAYTVVFGVVVVLVVSTLIPAASLPKT----
MAV_0729|M.avium_104 VPRPAMTPELAGHVSHHLSHAYTAVFVLAVVLVACTIIPASFLPRK----
MMAR_3100|M.marinum_M VPARAVGPDFLVELQHDLSHAYTLTFAVAAVLGALAYFPAAFLPAK----
Mflv_2221|M.gilvum_PYR-GCK LPQFLREPFALAMSQSMLLPAFVALFGVIAALFLFDLLGDRDAAAP--PA
Mvan_4474|M.vanbaalenii_PYR-1 LPAFLHAPFAAAMSQAMLLPAFVALFGVIAALFLVG-LGDRLPAPPRRPQ
MSMEG_5046|M.smegmatis_MC2_155 LPEFLREPFSAAMSQAMLLPAFIALFGVVAALFLLGFGSNRAAGHP----
TH_3965|M.thermoresistible__bu LPEFLHAPYAAAMAQSMLLPAFVALFGVLAAIFLAG----IVLSTP----
MAB_1396|M.abscessus_ATCC_1997 LPPGLRAPFSAAMAQSMLLGIAALALGLVAALFMRP-GGSQVGAVP----
MLBr_01562|M.leprae_Br4923 MNDAQLRALDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQE----
: * : ..:
Mb0805c|M.bovis_AF2122/97 ----------QAS-------HRRAPLLSA---------------------
Rv0783c|M.tuberculosis_H37Rv ----------QAS-------HRRAPLLSA---------------------
MUL_0501|M.ulcerans_Agy99 ----------PAAAPASFEQHHCDTQYGVHRV------------------
MAV_0729|M.avium_104 ----------PPS-----------PPAGD---------------------
MMAR_3100|M.marinum_M ----------PTT--------IAPGEVDVPG-------------------
Mflv_2221|M.gilvum_PYR-GCK PDVEPLEPFDPDHPLDWADGEDDYVEYEVSREE---FDRLATDG-TADGA
Mvan_4474|M.vanbaalenii_PYR-1 ETAEPVPAFDPDDELYWADGEDEYVEYEVPWDDGAEPRRPAQEADTVDAA
MSMEG_5046|M.smegmatis_MC2_155 ---------EPEHAGDYPD-DDEYVEIILSSDP-----------------
TH_3965|M.thermoresistible__bu ---------QPDADAQYLDEHDDYLVIELARPD-----------------
MAB_1396|M.abscessus_ATCC_1997 --------------------LDDVVSVED---------------------
MLBr_01562|M.leprae_Br4923 --------------------VKEAVDAGEL--------------------
Mb0805c|M.bovis_AF2122/97 --------------------------------------------------
Rv0783c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0501|M.ulcerans_Agy99 --------------------------------------------------
MAV_0729|M.avium_104 --------------------------------------------------
MMAR_3100|M.marinum_M --------------------------------------------------
Mflv_2221|M.gilvum_PYR-GCK AYN-ITEDFTEDITD-----VLDTVVEAPPAPADSTPSQQWRDLVDQLLD
Mvan_4474|M.vanbaalenii_PYR-1 ADTDVLDPVTEPMHDGGGYPAPESAPRRDHADPTSHPDREWRSILDQLLD
MSMEG_5046|M.smegmatis_MC2_155 ----------------------------PEDPPTDPQDDRWQHILDRLMQ
TH_3965|M.thermoresistible__bu ------------------------PNPRTDIAVTAVIPALRPTLRPTLRP
MAB_1396|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb0805c|M.bovis_AF2122/97 -----------------------------------
Rv0783c|M.tuberculosis_H37Rv -----------------------------------
MUL_0501|M.ulcerans_Agy99 -----------------------------------
MAV_0729|M.avium_104 -----------------------------------
MMAR_3100|M.marinum_M -----------------------------------
Mflv_2221|M.gilvum_PYR-GCK VSSPPGAP----RNGVGSADAEHPGGGPRV-----
Mvan_4474|M.vanbaalenii_PYR-1 PPKPSDDPTGQGRNGFHVTPSEDGRGASRGRHSLD
MSMEG_5046|M.smegmatis_MC2_155 -DAPPEDP-------------LPGRGPQRR-----
TH_3965|M.thermoresistible__bu SLGPPGGP----------VVQPNNTAALR------
MAB_1396|M.abscessus_ATCC_1997 -----------------------------------
MLBr_01562|M.leprae_Br4923 -----------------------------------