For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLDHIVLWTTDPRAAVDFYTQVVGLAPVRVSEFEAGDAPFPSVRVSADSIIDLLPRDSVADTETLTGGPG SAGHRVNHVCLAMTKAEYDALDQRLQAAGVDTSARLNRSYGARGWAPQGYYFADPDGNVVEARYYG*
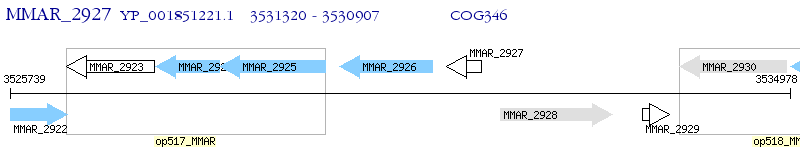
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2927 | - | - | 100% (137) | hypothetical protein MMAR_2927 |
| M. marinum M | MMAR_0534 | - | 2e-06 | 25.68% (148) | hypothetical protein MMAR_0534 |
| M. marinum M | MMAR_4165 | gloA_3 | 3e-05 | 32.59% (135) | glyoxalase, GloA_3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0280 | - | 7e-06 | 25.68% (148) | hypothetical protein Mb0280 |
| M. gilvum PYR-GCK | Mflv_0332 | - | 8e-06 | 25.00% (148) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0274 | - | 7e-06 | 25.68% (148) | hypothetical protein Rv0274 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4331c | - | 8e-05 | 25.34% (146) | putative glyoxalase/bleomycin resistance protein |
| M. avium 104 | MAV_0540 | - | 8e-54 | 69.12% (136) | dioxygenase |
| M. smegmatis MC2 155 | MSMEG_0608 | - | 2e-06 | 23.84% (151) | glyoxalase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1198 | - | 1e-06 | 25.68% (148) | hypothetical protein MUL_1198 |
| M. vanbaalenii PYR-1 | Mvan_0408 | - | 2e-06 | 27.03% (148) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4331c|M.abscessus_ATCC_199 MTMIKPNNPNSEFEFGGFNHVALVCSDMERTVDFYSNVLGMPLIKSLDLP
MSMEG_0608|M.smegmatis_MC2_155 --MIRPDNPNSEFEFGGINHVALVCSDMERTVDFYSNVLGMPLIKSLDLP
Mb0280|M.bovis_AF2122/97 --MIKPHNTNTEFELGGINHVALVCSDMARTVDFYSNILGMPLIKALDLP
Rv0274|M.tuberculosis_H37Rv --MIKPHNTNTEFELGGINHVALVCSDMARTVDFYSNILGMPLIKALDLP
MUL_1198|M.ulcerans_Agy99 --MIKPHNTNTEFEFGGINHVALVCSDMARTVDFYSNILGMPLIKSLDLP
Mflv_0332|M.gilvum_PYR-GCK --MIKPDNTNTEFALGGINHVALVCSDMARTVDFYSNVLGMPLIKSLDLP
Mvan_0408|M.vanbaalenii_PYR-1 --MIKPQNTNTEFELGGINHVALVCSDMGKTVDFYTNVLGMPLVKSLDLP
MMAR_2927|M.marinum_M ---------------MRLDHIVLWTTDPRAAVDFYTQVVGLAPVRVSEFE
MAV_0540|M.avium_104 ---------------MRLDHVVLWTRNPRVSMDFYTRVVGLEPVRFAEFE
::*:.* : ::***:.::*: :: ::
MAB_4331c|M.abscessus_ATCC_199 MGQGQHFFFDAGGGDSLAFFWFKDAPDGVPGISAPAAIPGIGDIVSAVSS
MSMEG_0608|M.smegmatis_MC2_155 GGIGQHFFFDAGNGDCIAFFWFRDAPDGVPGLTAPAAIPGIGEIVSATGS
Mb0280|M.bovis_AF2122/97 GGQGQHFFFDAGNGDCVAFFWFADAPDRVPGLSSPVAIPGIGDITSAVST
Rv0274|M.tuberculosis_H37Rv GGQGQHFFFDAGNGDCVAFFWFADAPDRVPGLSSPVAIPGIGDITSAVST
MUL_1198|M.ulcerans_Agy99 GGQDQHFFFDAGNGDCVAFFWFADAPDRVPGISSPGAIPGIGDITSAVST
Mflv_0332|M.gilvum_PYR-GCK GGMGQHFFFDAGNGDCVAFFWFAEAPDRVPGISSPEAIPGIGDIVSAVST
Mvan_0408|M.vanbaalenii_PYR-1 GGMGQHFFFDAGNGDCVAFFWFADAPDRVPGISSPEAIPGIGDIVSAVST
MMAR_2927|M.marinum_M AGDAPFPSVRVSADSIIDLLPR----DSVADTETLTGGPGS-----AGHR
MAV_0540|M.avium_104 AGEAPFPSVRICADSIIDLMPL----EMVSGAASSGVAEGS-----AGNP
* . . .. : :: : *.. : * *
MAB_4331c|M.abscessus_ATCC_199 MNHISLHVPAEKFDEYRVKLKAKGVRVGPILNHDESEFQVSRELHPGVYV
MSMEG_0608|M.smegmatis_MC2_155 LNHIALHVPAEKFDEYREKLKAKGVRVGPILNHDNSEMQVSPTVHPGVYV
Mb0280|M.bovis_AF2122/97 MNHLAFHVPAERFDAYRQRLKDKGVRVGPVLNHDDSETQVSAVVHPGVYV
Rv0274|M.tuberculosis_H37Rv MNHLAFHVPAERFDAYRQRLKDKGVRVGPVLNHDDSETQVSAVVHPGVYV
MUL_1198|M.ulcerans_Agy99 MNHLAFHVPAEKFDAYRQRLKDKGVRVGPVLNHDHSEFQVSATVHPGVYV
Mflv_0332|M.gilvum_PYR-GCK MNHLAFHVPAEKFDEYRQKLKAKGVRVGPVLNHDESPAQVSATLHPGVYV
Mvan_0408|M.vanbaalenii_PYR-1 MNHLAFHVPAEKFDEYRQRLKDKGVRVGPVLNHDESEAQVSATVHPGVYV
MMAR_2927|M.marinum_M VNHVCLAMTKAEYDALDQRLQAAGVDTSARLNRSYG--------ARGWAP
MAV_0540|M.avium_104 VNHVCIALSKAEYDALEARLQAEGVDTGVRLSRSFG--------ARGWSP
:**:.: :. .:* :*: ** .. *.:. . *
MAB_4331c|M.abscessus_ATCC_199 RSFYFLDPDGITLEFACWTKEFTAADVAVAPKTAAERTPREVATA-----
MSMEG_0608|M.smegmatis_MC2_155 RSFYFFDPDGITLEFACWTKEFTDADTHAVPKTAADRRPKTAQPGLVAST
Mb0280|M.bovis_AF2122/97 RSFYFQDPDGITLEFACWTKEFTTSDAQAVPKTAADRRPPVAADR-----
Rv0274|M.tuberculosis_H37Rv RSFYFQDPDGITLEFACWTKEFTTSDAQAVPKTAADRRPPVAADR-----
MUL_1198|M.ulcerans_Agy99 RSFYFQDPDGITLEFACWIKEFTADHTPAVPRTAADRRPPVAAGH-----
Mflv_0332|M.gilvum_PYR-GCK RSFYFLDPDGITLEFACWTKEFGDSDTATVPKTAADRRVPVNG-------
Mvan_0408|M.vanbaalenii_PYR-1 RSFYFLDPDGITLEFACWTKQFGDTDTTTVPKTEADRRVQVSG-------
MMAR_2927|M.marinum_M QGYYFADPDGNVVEARYYG-------------------------------
MAV_0540|M.avium_104 HTYYFADPDGNVVEARYYE-------------------------------
: :** **** .:* :
MAB_4331c|M.abscessus_ATCC_199 -
MSMEG_0608|M.smegmatis_MC2_155 K
Mb0280|M.bovis_AF2122/97 -
Rv0274|M.tuberculosis_H37Rv -
MUL_1198|M.ulcerans_Agy99 -
Mflv_0332|M.gilvum_PYR-GCK -
Mvan_0408|M.vanbaalenii_PYR-1 -
MMAR_2927|M.marinum_M -
MAV_0540|M.avium_104 -