For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIRPDNPNSEFEFGGINHVALVCSDMERTVDFYSNVLGMPLIKSLDLPGGIGQHFFFDAGNGDCIAFFWF RDAPDGVPGLTAPAAIPGIGEIVSATGSLNHIALHVPAEKFDEYREKLKAKGVRVGPILNHDNSEMQVSP TVHPGVYVRSFYFFDPDGITLEFACWTKEFTDADTHAVPKTAADRRPKTAQPGLVASTK
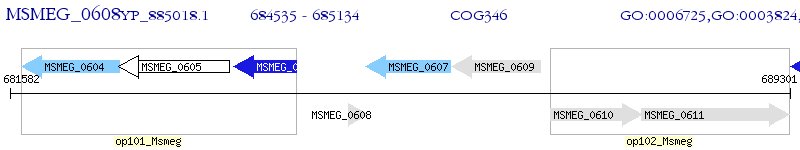
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0608 | - | - | 100% (199) | glyoxalase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0280 | - | 3e-91 | 78.95% (190) | hypothetical protein Mb0280 |
| M. gilvum PYR-GCK | Mflv_0332 | - | 2e-92 | 81.72% (186) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0274 | - | 3e-91 | 78.95% (190) | hypothetical protein Rv0274 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4331c | - | 2e-93 | 83.51% (188) | putative glyoxalase/bleomycin resistance protein |
| M. marinum M | MMAR_0534 | - | 1e-91 | 80.00% (190) | hypothetical protein MMAR_0534 |
| M. avium 104 | MAV_4885 | - | 2e-90 | 78.72% (188) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. thermoresistible (build 8) | TH_0800 | - | 5e-93 | 81.28% (187) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1198 | - | 3e-90 | 78.95% (190) | hypothetical protein MUL_1198 |
| M. vanbaalenii PYR-1 | Mvan_0408 | - | 8e-91 | 79.57% (186) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0280|M.bovis_AF2122/97 --MIKPHNTNTEFELGGINHVALVCSDMARTVDFYSNILGMPLIKALDLP
Rv0274|M.tuberculosis_H37Rv --MIKPHNTNTEFELGGINHVALVCSDMARTVDFYSNILGMPLIKALDLP
MMAR_0534|M.marinum_M --MIKPHNTNTEFEFGGINHVALVCSDMARTVDFYSNILGMPLIKSLDLP
MUL_1198|M.ulcerans_Agy99 --MIKPHNTNTEFEFGGINHVALVCSDMARTVDFYSNILGMPLIKSLDLP
MAV_4885|M.avium_104 --MIKPHNTNTEFELGGINHVALVCSDMAKTVDFYSNVLGMPLVKSLDLP
Mflv_0332|M.gilvum_PYR-GCK --MIKPDNTNTEFALGGINHVALVCSDMARTVDFYSNVLGMPLIKSLDLP
Mvan_0408|M.vanbaalenii_PYR-1 --MIKPQNTNTEFELGGINHVALVCSDMGKTVDFYTNVLGMPLVKSLDLP
TH_0800|M.thermoresistible__bu --MIKPRNRNPEFELGGINHVALVCSDMAKTVDFYGNVLGMPLVKSLDLP
MSMEG_0608|M.smegmatis_MC2_155 --MIRPDNPNSEFEFGGINHVALVCSDMERTVDFYSNVLGMPLIKSLDLP
MAB_4331c|M.abscessus_ATCC_199 MTMIKPNNPNSEFEFGGFNHVALVCSDMERTVDFYSNVLGMPLIKSLDLP
**:* * *.** :**:********** :***** *:*****:*:****
Mb0280|M.bovis_AF2122/97 GGQGQHFFFDAGNGDCVAFFWFADAPDRVPGLSSPVAIPGIGDITSAVST
Rv0274|M.tuberculosis_H37Rv GGQGQHFFFDAGNGDCVAFFWFADAPDRVPGLSSPVAIPGIGDITSAVST
MMAR_0534|M.marinum_M GGQGQHFFFDAGNGDCVAFFWFADAPDRVPGISSPGAIPGIGDITSAVST
MUL_1198|M.ulcerans_Agy99 GGQDQHFFFDAGNGDCVAFFWFADAPDRVPGISSPGAIPGIGDITSAVST
MAV_4885|M.avium_104 GGAGQHFFFDAGNGDCVAFFWFADAPDRVPGISSPGAIPGIGDITSAVST
Mflv_0332|M.gilvum_PYR-GCK GGMGQHFFFDAGNGDCVAFFWFAEAPDRVPGISSPEAIPGIGDIVSAVST
Mvan_0408|M.vanbaalenii_PYR-1 GGMGQHFFFDAGNGDCVAFFWFADAPDRVPGISSPEAIPGIGDIVSAVST
TH_0800|M.thermoresistible__bu GGMGQHFFFDCGNGDCIAFFWFRDAPDGVPGISAPRHIPGTGDIVSAVSS
MSMEG_0608|M.smegmatis_MC2_155 GGIGQHFFFDAGNGDCIAFFWFRDAPDGVPGLTAPAAIPGIGEIVSATGS
MAB_4331c|M.abscessus_ATCC_199 MGQGQHFFFDAGGGDSLAFFWFKDAPDGVPGISAPAAIPGIGDIVSAVSS
* .******.*.**.:***** :*** ***:::* *** *:*.**..:
Mb0280|M.bovis_AF2122/97 MNHLAFHVPAERFDAYRQRLKDKGVRVGPVLNHDDSETQVSAVVHPGVYV
Rv0274|M.tuberculosis_H37Rv MNHLAFHVPAERFDAYRQRLKDKGVRVGPVLNHDDSETQVSAVVHPGVYV
MMAR_0534|M.marinum_M MNHLAFHVPAEKFDAYRQRLKDKGVRVGPVLNHDHSEFQVSATVHPGVYV
MUL_1198|M.ulcerans_Agy99 MNHLAFHVPAEKFDAYRQRLKDKGVRVGPVLNHDHSEFQVSATVHPGVYV
MAV_4885|M.avium_104 MNHLAFHVPAEKFDAYRQRLKDKGVRVGPVLNHDESPMQVSATVHPGVYV
Mflv_0332|M.gilvum_PYR-GCK MNHLAFHVPAEKFDEYRQKLKAKGVRVGPVLNHDESPAQVSATLHPGVYV
Mvan_0408|M.vanbaalenii_PYR-1 MNHLAFHVPAEKFDEYRQRLKDKGVRVGPVLNHDESEAQVSATVHPGVYV
TH_0800|M.thermoresistible__bu MNHLAFHVPAEKFDEYRRRLKEKGVRVGPVLNHDDSETQVSPTVHPGVYV
MSMEG_0608|M.smegmatis_MC2_155 LNHIALHVPAEKFDEYREKLKAKGVRVGPILNHDNSEMQVSPTVHPGVYV
MAB_4331c|M.abscessus_ATCC_199 MNHISLHVPAEKFDEYRVKLKAKGVRVGPILNHDESEFQVSRELHPGVYV
:**:::*****:** ** :** *******:****.* *** :******
Mb0280|M.bovis_AF2122/97 RSFYFQDPDGITLEFACWTKEFTTSDAQAVPKTAADRRPPVAADR-----
Rv0274|M.tuberculosis_H37Rv RSFYFQDPDGITLEFACWTKEFTTSDAQAVPKTAADRRPPVAADR-----
MMAR_0534|M.marinum_M RSFYFQDPDGITLEFACWIKEFTADDTPAVPRTAADRRPPVAAGH-----
MUL_1198|M.ulcerans_Agy99 RSFYFQDPDGITLEFACWIKEFTADHTPAVPRTAADRRPPVAAGH-----
MAV_4885|M.avium_104 RSFYFQDPDGITLEFACWVKEFDSGDARAVPKTAADRRPRVGAGR-----
Mflv_0332|M.gilvum_PYR-GCK RSFYFLDPDGITLEFACWTKEFGDSDTATVPKTAADRRVPVNG-------
Mvan_0408|M.vanbaalenii_PYR-1 RSFYFLDPDGITLEFACWTKQFGDTDTTTVPKTEADRRVQVSG-------
TH_0800|M.thermoresistible__bu RSFYFHDPDGITLEFACWTKEFTDTDAQVSPRTAADRRPSVTA-------
MSMEG_0608|M.smegmatis_MC2_155 RSFYFFDPDGITLEFACWTKEFTDADTHAVPKTAADRRPKTAQPGLVAST
MAB_4331c|M.abscessus_ATCC_199 RSFYFLDPDGITLEFACWTKEFTAADVAVAPKTAAERTPREVATA-----
***** ************ *:* .. . *:* *:*
Mb0280|M.bovis_AF2122/97 -
Rv0274|M.tuberculosis_H37Rv -
MMAR_0534|M.marinum_M -
MUL_1198|M.ulcerans_Agy99 -
MAV_4885|M.avium_104 -
Mflv_0332|M.gilvum_PYR-GCK -
Mvan_0408|M.vanbaalenii_PYR-1 -
TH_0800|M.thermoresistible__bu -
MSMEG_0608|M.smegmatis_MC2_155 K
MAB_4331c|M.abscessus_ATCC_199 -