For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDPVSVIARFAPLPERRADLRVLLAGMVGPTRSEEGCRRYDLYEAPPDGELVLIERYDDQAALEHHRTT EHYLNYRAQLSALLASPITVSVLAPLDEAEASRHRLRE
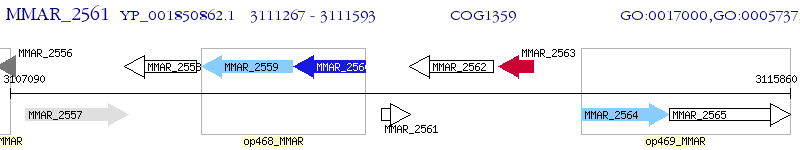
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2561 | - | - | 100% (108) | hypothetical protein MMAR_2561 |
| M. marinum M | MMAR_1966 | - | 8e-05 | 28.16% (103) | hypothetical protein MMAR_1966 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0817 | - | 6e-31 | 59.41% (101) | hypothetical protein Mb0817 |
| M. gilvum PYR-GCK | Mflv_3995 | - | 1e-05 | 25.00% (104) | antibiotic biosynthesis monooxygenase |
| M. tuberculosis H37Rv | Rv0793 | - | 6e-31 | 59.41% (101) | hypothetical protein Rv0793 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3640 | - | 9e-05 | 26.67% (105) | antibiotic biosynthesis monooxygenase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_2688 | - | 8e-05 | 27.88% (104) | antibiotic biosynthesis monooxygenase domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3200 | - | 4e-57 | 99.07% (108) | hypothetical protein MUL_3200 |
| M. vanbaalenii PYR-1 | Mvan_2392 | - | 1e-06 | 29.81% (104) | antibiotic biosynthesis monooxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3995|M.gilvum_PYR-GCK --MPIVVVATLTAKPESVETVRNACKQAIEAVHEEPGCQLYSLHEA--DN
Mvan_2392|M.vanbaalenii_PYR-1 --MPVVVIATMKAKPESVDAVRNACQTAIESVHDEPGCDLYALHEA--DG
MSMEG_2688|M.smegmatis_MC2_155 --MPVVVVATMKAKPESVDAVREECTKAVAAVHDEPGCELYALHEH--DG
MAV_3640|M.avium_104 --MPVVVVATMTVKPESVDTVRDILTRAVEEVHDEPGCQLYSLHQS--GE
MMAR_2561|M.marinum_M MSDPVSVIARFAPLPERRADLRVLLAGMVGPTRSEEGCRRYDLYEAPPDG
MUL_3200|M.ulcerans_Agy99 MSDPVSVIARFAPLPERRADLRVLLAGMVGPTRSEEGCRRYDLYEAPPDG
Mb0817|M.bovis_AF2122/97 MTSPVAVIARFMPRPDARSALRALLDAMITPTRAEDGCRSYDLYESADGG
Rv0793|M.tuberculosis_H37Rv MTSPVAVIARFMPRPDARSALRALLDAMITPTRAEDGCRSYDLYESADGG
*: *:* : *: :* : .: * ** * *:: .
Mflv_3995|M.gilvum_PYR-GCK TFVFVEQWEDADALQKHSAAPAVATMFGAVGEHLDGAPDIKVLQPVVAGD
Mvan_2392|M.vanbaalenii_PYR-1 TFVFVEQWADADALQKHSTAPAVATLFGTVGELLDGAPDIKMLQPMVAGD
MSMEG_2688|M.smegmatis_MC2_155 TFVFVEQWADADALKTHGGAPAIGALFGAIGSLLDGAPDIKTLQPVVAGD
MAV_3640|M.avium_104 TFVFVEQWADEEALKTHSTAPAIGKMFSAAGEHLDGAPDIKMLQPVPAGD
MMAR_2561|M.marinum_M ELVLIERYDDQAALEHHRTTEHYLNYRAQLSALLASPITVSVLAPLDEAE
MUL_3200|M.ulcerans_Agy99 ELVLIERYDDQAALEHHRTTEDYLNYRAQLSALLASPITVSVLAPLDEAE
Mb0817|M.bovis_AF2122/97 ELVLFERYRSRIALDEHRGSPHYLNYRAQVGELLTRPVAVTVLAPLDEAS
Rv0793|M.tuberculosis_H37Rv ELVLFERYRSRIALDEHRGSPHYLNYRAQVGELLTRPVAVTVLAPLDEAS
:*:.*:: . **. * : . . * . :. * *: ..
Mflv_3995|M.gilvum_PYR-GCK PAKGQLRA
Mvan_2392|M.vanbaalenii_PYR-1 PAKGRLRP
MSMEG_2688|M.smegmatis_MC2_155 PAKGRLRP
MAV_3640|M.avium_104 PGKGQLRP
MMAR_2561|M.marinum_M ASRHRLRE
MUL_3200|M.ulcerans_Agy99 ASRHRLRE
Mb0817|M.bovis_AF2122/97 A-------
Rv0793|M.tuberculosis_H37Rv A-------
.