For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVVVATLTVKPESVDAVRDILTTAVEEVHGEPGCQLYSLHQTGETFVFIEQWADAEALTAHSTAPAVAKM FTAAGEHLAGAPDIKMLAPIPAGDPDKGQLRQ*
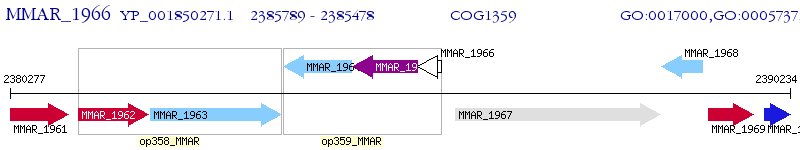
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1966 | - | - | 100% (103) | hypothetical protein MMAR_1966 |
| M. marinum M | MMAR_2133 | - | 3e-05 | 36.00% (75) | hypothetical protein MMAR_2133 |
| M. marinum M | MMAR_2561 | - | 7e-05 | 28.16% (103) | hypothetical protein MMAR_2561 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2770 | - | 1e-45 | 81.19% (101) | hypothetical protein Mb2770 |
| M. gilvum PYR-GCK | Mflv_3995 | - | 1e-36 | 70.00% (100) | antibiotic biosynthesis monooxygenase |
| M. tuberculosis H37Rv | Rv2749 | - | 1e-45 | 81.19% (101) | hypothetical protein Rv2749 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3080 | - | 5e-23 | 48.51% (101) | putative antibiotic biosynthesis monooxygenase |
| M. avium 104 | MAV_3640 | - | 3e-46 | 84.00% (100) | antibiotic biosynthesis monooxygenase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_2688 | - | 6e-32 | 63.00% (100) | antibiotic biosynthesis monooxygenase domain-containing protein |
| M. thermoresistible (build 8) | TH_0832 | - | 8e-38 | 72.00% (100) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3391 | - | 5e-54 | 99.01% (101) | hypothetical protein MUL_3391 |
| M. vanbaalenii PYR-1 | Mvan_2392 | - | 3e-35 | 67.96% (103) | antibiotic biosynthesis monooxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1966|M.marinum_M MP-VVVATLTVKPESVDAVRDILTTAVEEVHGEPGCQLYSLHQTGETFVF
MUL_3391|M.ulcerans_Agy99 MPVVVVATLTVKPESVDAVRDILTTAVEEVHGEPGCQLYSLHQTGETFVF
MAV_3640|M.avium_104 MPVVVVATMTVKPESVDTVRDILTRAVEEVHDEPGCQLYSLHQSGETFVF
Mb2770|M.bovis_AF2122/97 MPVVVVATLTAKPESVDTVRDILTRAVDDVHREPGCQLYALHETGETFIF
Rv2749|M.tuberculosis_H37Rv MPVVVVATLTAKPESVDTVRDILTRAVDDVHREPGCQLYALHETGETFIF
TH_0832|M.thermoresistible__bu MPVVVVASLTAKPESVDTVREACKRAVEAVHKEPGCDLYALHQTDRTFVF
Mflv_3995|M.gilvum_PYR-GCK MPIVVVATLTAKPESVETVRNACKQAIEAVHEEPGCQLYSLHEADNTFVF
MSMEG_2688|M.smegmatis_MC2_155 MPVVVVATMKAKPESVDAVREECTKAVAAVHDEPGCELYALHEHDGTFVF
Mvan_2392|M.vanbaalenii_PYR-1 MPVVVIATMKAKPESVDAVRNACQTAIESVHDEPGCDLYALHEADGTFVF
MAB_3080|M.abscessus_ATCC_1997 MP-TVVAFLYPQPDKRDEVRAALLETIPLVHDEPGCNLYTLHEAQDRFVF
** .*:* : :*:. : ** :: ** ****:**:**: *:*
MMAR_1966|M.marinum_M IEQWADAEALTAHSTAPAVAKMFTAAGEHLAGAPDIKMLAPIPAGDPDKG
MUL_3391|M.ulcerans_Agy99 IEQWADAEALTAHSTAPAVAKMFTAAGEHLAGAPNIKMLAPIPAGDPDKG
MAV_3640|M.avium_104 VEQWADEEALKTHSTAPAIGKMFSAAGEHLDGAPDIKMLQPVPAGDPGKG
Mb2770|M.bovis_AF2122/97 VEQWADAEALKAHSGAPAVATMFTAAGEHLVGAPDIKLLQPVPAGDPSKG
Rv2749|M.tuberculosis_H37Rv VEQWADAEALKAHSGAPAVATMFTAAGEHLVGAPDIKLLQPVPAGDPSKG
TH_0832|M.thermoresistible__bu IEQWADEEALRTHSAAPAVTTMFAEIGEHLDGAPDIKMLQPVPAGDPAKG
Mflv_3995|M.gilvum_PYR-GCK VEQWEDADALQKHSAAPAVATMFGAVGEHLDGAPDIKVLQPVVAGDPAKG
MSMEG_2688|M.smegmatis_MC2_155 VEQWADADALKTHGGAPAIGALFGAIGSLLDGAPDIKTLQPVVAGDPAKG
Mvan_2392|M.vanbaalenii_PYR-1 VEQWADADALQKHSTAPAVATLFGTVGELLDGAPDIKMLQPMVAGDPAKG
MAB_3080|M.abscessus_ATCC_1997 VESWASGEALAEHGKAPAVTAMFGKVGPLLSQPPEIVVAEALPAGDPAKG
:*.* . :** *. ***: :* * * .*:* .: **** **
MMAR_1966|M.marinum_M QLRQ
MUL_3391|M.ulcerans_Agy99 QLRQ
MAV_3640|M.avium_104 QLRP
Mb2770|M.bovis_AF2122/97 QLRR
Rv2749|M.tuberculosis_H37Rv QLRR
TH_0832|M.thermoresistible__bu QVRG
Mflv_3995|M.gilvum_PYR-GCK QLRA
MSMEG_2688|M.smegmatis_MC2_155 RLRP
Mvan_2392|M.vanbaalenii_PYR-1 RLRP
MAB_3080|M.abscessus_ATCC_1997 DLVH
: