For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQILPTQGASGHAGRSHFGPTDRKLWVLACMDERLPINEALGIQVDSPVGHGDAHCFRNAGGLVTDDAIR SAMLSTNFFGTTEIVIVQHTQCGMLSANANDLEAYFKAKGIDTDEPILDPTLPELKLDKGGFAKWIGMTD DVDEALLKTIEVFENHPLIPDDVTISGWVWEVETRRLRPPTRDPARRARTDATARDYGITQPQPARWHLS DGGKNVATATDPVCGMELVVDCAAGYREADGYRQYFCSQVCLDSYDADPARYQPVAHTM*
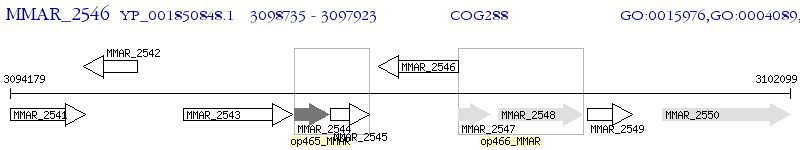
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2546 | - | - | 100% (270) | hypothetical protein MMAR_2546 |
| M. marinum M | MMAR_1420 | cynT | e-138 | 92.37% (249) | carbonic anhydrase, CynT |
| M. marinum M | MMAR_4135 | - | 4e-20 | 34.57% (162) | carbonic anhydrase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1315 | - | 6e-17 | 34.36% (163) | hypothetical protein Mb1315 |
| M. gilvum PYR-GCK | Mflv_2270 | - | 2e-18 | 33.13% (163) | carbonic anhydrase |
| M. tuberculosis H37Rv | Rv1284 | - | 7e-17 | 34.36% (163) | hypothetical protein Rv1284 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4126c | - | 1e-16 | 32.10% (162) | carbonic anhydrase-related protein |
| M. avium 104 | MAV_1434 | - | 8e-19 | 32.91% (158) | carbonic anhydrase |
| M. smegmatis MC2 155 | MSMEG_4985 | - | 1e-16 | 28.83% (163) | carbonic anhydrase |
| M. thermoresistible (build 8) | TH_1318 | - | 3e-19 | 34.76% (164) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3998 | - | 3e-20 | 34.57% (162) | carbonic anhydrase |
| M. vanbaalenii PYR-1 | Mvan_4424 | - | 5e-17 | 31.90% (163) | carbonic anhydrase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2270|M.gilvum_PYR-GCK -MTVTDQYLKNNEKYAETFS-GPLPLPPSKHVAVVACMDARLDVYRILGL
Mvan_4424|M.vanbaalenii_PYR-1 -MSVTDEYLKNNEEYAKTFS-GPLPLPPSRHVAVVACMDARLDVYRILGL
MSMEG_4985|M.smegmatis_MC2_155 -MSVTDQYLANNEEYAKTFS-GPLPLPPSKHVAVVACMDARLDVYRILGL
TH_1318|M.thermoresistible__bu -MSVTDEYLRNNEEYAKTFK-GPLPLPPSKHVAIVACMDARLDVYRALGI
Mb1315|M.bovis_AF2122/97 -MTVTDDYLANNVDYASGFK-GPLPMPPSKHIAIVACMDARLDVYRMLGI
Rv1284|M.tuberculosis_H37Rv -MTVTDDYLANNVDYASGFK-GPLPMPPSKHIAIVACMDARLDVYRMLGI
MUL_3998|M.ulcerans_Agy99 -MTVTDDYLANNADYASGFE-GPLPMPPSKHIAILACMDARLDVYRLLGI
MAV_1434|M.avium_104 -MAVTDDYLAHNAGYARSFE-GPLPMPPSKHVAVVACMDARLDVYRILGL
MAB_4126c|M.abscessus_ATCC_199 -MSTTDELLKNAQAYAASFDKGELPLPPARKVAVVACMDARLNPYGLLGL
MMAR_2546|M.marinum_M MTQILPTQGASGHAGRSHFG------PTDRKLWVLACMDERLPINEALGI
* *. ::: ::**** ** **:
Mflv_2270|M.gilvum_PYR-GCK GD------GEAHVIRNAGGVVTDDEIRSLAISQRLLGTREIILIHHTDCG
Mvan_4424|M.vanbaalenii_PYR-1 KD------GEAHVIRNAGGVVTDDEIRSLAISQRLLGTREIILIHHTDCG
MSMEG_4985|M.smegmatis_MC2_155 GD------GEAHVIRNAGGVITDDEIRSLAISQRLLGTKEIILIHHTDCG
TH_1318|M.thermoresistible__bu NE------GEAHVIRNAGGVITDDEIRSLAISQRLLGTREIILIHHTDCG
Mb1315|M.bovis_AF2122/97 KE------GEAHVIRNAGCVVTDDVIRSLAISQRLLGTREIILLHHTDCG
Rv1284|M.tuberculosis_H37Rv KE------GEAHVIRNAGCVVTDDVIRSLAISQRLLGTREIILLHHTDCG
MUL_3998|M.ulcerans_Agy99 NE------GEAHVIRNAGGVATDDAIRSLAISQRLLGTREVVLIHHTDCG
MAV_1434|M.avium_104 RE------GEAHVIRNAGGVITDDVVRSLAISQRLLGTREIILIHHTDCG
MAB_4126c|M.abscessus_ATCC_199 HE------GDAHVIRNAGGVITEDEIRSLAISQRLLGTEEIILIHHTDCG
MMAR_2546|M.marinum_M QVDSPVGHGDAHCFRNAGGLVTDDAIRSAMLSTNFFGTTEIVIVQHTQCG
*:** :**** : *:* :** :* .::** *:::::**:**
Mflv_2270|M.gilvum_PYR-GCK MLTFSDDGFK-----QQIQDETGLKPEWAAEAFQD-------------VE
Mvan_4424|M.vanbaalenii_PYR-1 MLTFTDDGFK-----QQIQDEIGIKPEWAAEAFQD-------------PD
MSMEG_4985|M.smegmatis_MC2_155 MLTFTDDEFK-----RAIQGETGIKPEWAAESFTD-------------LE
TH_1318|M.thermoresistible__bu MLTFTDDDFK-----KQIEEEIGIRPPWAPESFSD-------------PA
Mb1315|M.bovis_AF2122/97 MLTFTDDDFK-----RAIQDETGIRPTWSPESYPD-------------AV
Rv1284|M.tuberculosis_H37Rv MLTFTDDDFK-----RAIQDETGIRPTWSPESYPD-------------AV
MUL_3998|M.ulcerans_Agy99 MLTFTDDDFK-----RGIQDETGVKPPWAAEAFPD-------------AA
MAV_1434|M.avium_104 MLTFTDDDFK-----RGIQEETGIKPPWAAEAFAD-------------LA
MAB_4126c|M.abscessus_ATCC_199 MLTFTDDAFK-----QSIQDETGIKPPWSAEAFTD-------------LD
MMAR_2546|M.marinum_M MLSANANDLEAYFKAKGIDTDEPILDPTLPELKLDKGGFAKWIGMTDDVD
**: . : :: : *: : : .* *
Mflv_2270|M.gilvum_PYR-GCK EDVRQSLRRIEASPFVTKHESLRGFVFDVTTGRLNEVTL-----------
Mvan_4424|M.vanbaalenii_PYR-1 EDVRQSLRRIDASPFVTKHESLRGFVFDVATGRLNEVTP-----------
MSMEG_4985|M.smegmatis_MC2_155 EDVRQSLRRIEASPFVTKHESLRGFIFDVATGKLAEVTL-----------
TH_1318|M.thermoresistible__bu EDVRQSLRRIEASPFVTLHESLRGFVFDVATGRLTEVTLD----------
Mb1315|M.bovis_AF2122/97 EDVRQSLRRIEVNPFVTKHTSLRGFVFDVATGKLNEVTP-----------
Rv1284|M.tuberculosis_H37Rv EDVRQSLRRIEVNPFVTKHTSLRGFVFDVATGKLNEVTP-----------
MUL_3998|M.ulcerans_Agy99 EDVKQSLRRIENSPFVTKHVSLRGFVFDVATGKLNEVTL-----------
MAV_1434|M.avium_104 EDVRQSLRRIEANPFVTKHVSARGFVFDVATGKLDEVKP-----------
MAB_4126c|M.abscessus_ATCC_199 EDVRQSLARINANPFVPRKDSVRGFVYDVTNGTLREVTAAA---------
MMAR_2546|M.marinum_M EALLKTIEVFENHPLIPDDVTISGWVWEVETRRLRPPTRDPARRARTDAT
* : ::: :: *::. . : *::::* . * .
Mflv_2270|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4424|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4985|M.smegmatis_MC2_155 --------------------------------------------------
TH_1318|M.thermoresistible__bu --------------------------------------------------
Mb1315|M.bovis_AF2122/97 --------------------------------------------------
Rv1284|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3998|M.ulcerans_Agy99 --------------------------------------------------
MAV_1434|M.avium_104 --------------------------------------------------
MAB_4126c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2546|M.marinum_M ARDYGITQPQPARWHLSDGGKNVATATDPVCGMELVVDCAAGYREADGYR
Mflv_2270|M.gilvum_PYR-GCK --------------------------
Mvan_4424|M.vanbaalenii_PYR-1 --------------------------
MSMEG_4985|M.smegmatis_MC2_155 --------------------------
TH_1318|M.thermoresistible__bu --------------------------
Mb1315|M.bovis_AF2122/97 --------------------------
Rv1284|M.tuberculosis_H37Rv --------------------------
MUL_3998|M.ulcerans_Agy99 --------------------------
MAV_1434|M.avium_104 --------------------------
MAB_4126c|M.abscessus_ATCC_199 --------------------------
MMAR_2546|M.marinum_M QYFCSQVCLDSYDADPARYQPVAHTM