For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RILPETTSVTEYYGHKRPEMLRFLPADAQCVLELGCGEGVFGAIVKQSTGAEVWGLEYNEEAAQRAGELL DHVLAGDASTRIAELPDNYFDAVVCNDVLEHLVDPLDTLKRLRPKLKPDGVVVASIPNIRYLPALGKVVF RRDFPQEDFGIFDRTHLRFFTRSSMVRMFEDAGFSMQRIKGINPFLGPFGALFVALSLGHFADGVFLQYA CVATPAS*
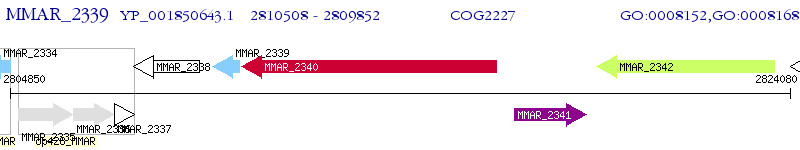
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2339 | - | - | 100% (218) | SAM-dependent methyltransferase |
| M. marinum M | MMAR_2350 | - | 2e-74 | 58.72% (218) | methylase |
| M. marinum M | MMAR_1683 | - | 1e-10 | 30.22% (182) | hypothetical protein MMAR_1683 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3056 | - | 5e-09 | 29.67% (182) | hypothetical protein Mb3056 |
| M. gilvum PYR-GCK | Mflv_4260 | - | 3e-09 | 30.82% (159) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3030 | - | 5e-09 | 29.67% (182) | hypothetical protein Rv3030 |
| M. leprae Br4923 | MLBr_01713 | - | 2e-06 | 27.22% (158) | hypothetical protein MLBr_01713 |
| M. abscessus ATCC 19977 | MAB_3364 | - | 1e-09 | 30.19% (159) | hypothetical protein MAB_3364 |
| M. avium 104 | MAV_3877 | - | 1e-08 | 30.19% (159) | hypothetical protein MAV_3877 |
| M. smegmatis MC2 155 | MSMEG_2350 | - | 4e-10 | 30.72% (153) | hypothetical protein MSMEG_2350 |
| M. thermoresistible (build 8) | TH_2193 | - | 4e-08 | 28.93% (159) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1921 | - | 7e-11 | 30.22% (182) | hypothetical protein MUL_1921 |
| M. vanbaalenii PYR-1 | Mvan_2098 | - | 2e-10 | 30.72% (153) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2350|M.smegmatis_MC2_155 ----------------MGDPDNALPSALPLTGERTIPGLAEENYWFRRHE
TH_2193|M.thermoresistible__bu ----------MSAFSTAGSDADGPISVLPLTGERTVPGLERENYWFRRHE
Mflv_4260|M.gilvum_PYR-GCK ----------MSASVTNG---DPG---LPLTGERTIPGLAEENYWFRRHE
Mvan_2098|M.vanbaalenii_PYR-1 ----------MSASVTDGPTEDPAP--MALTGERTVPGVAEENYWFRRHE
MAB_3364|M.abscessus_ATCC_1997 MNSHLTDPHPADPLRTSGPSAEDPDATLALTGERTVPGIAEENYWFRRHE
Mb3056|M.bovis_AF2122/97 MCAFVPHVPRHSRGDNPPSASTASPAVLTLTGERTIPDLDIENYWFRRHQ
Rv3030|M.tuberculosis_H37Rv MCAFVPHVPRHSRGDNPPSASTASPAVLTLTGERTIPDLDIENYWFRRHQ
MLBr_01713|M.leprae_Br4923 MSAFVPDVPNRIPDDPSSIDSTAFHGMLTLTGERTIPDLDIENYWFRRHQ
MUL_1921|M.ulcerans_Agy99 MSSFVPDVPDQIP----TASAPPVDAVLTLTGERTIPDLDIENYWFRRHE
MAV_3877|M.avium_104 MSALVPDLPPHPP----AGAAPAGDAVMMLTGERTIPGLDIENYWFRRHE
MMAR_2339|M.marinum_M --------------------------------MRILPETTSVTEYYGHKR
* :* . :: ::.
MSMEG_2350|M.smegmatis_MC2_155 VVYQRLAHRCAGRDVLEAGCGEG-YGADLIADVARRVIGLDYDEATVAHV
TH_2193|M.thermoresistible__bu VVYQRLAHRCAGRVVLEAGCGEG-YGADLLAEVAAHVIGLDYDAATVAHV
Mflv_4260|M.gilvum_PYR-GCK VVYRRLVERCRDRDVLEAGSGEG-YGADLIAEVARSVIGLDYDESAVAHV
Mvan_2098|M.vanbaalenii_PYR-1 VVYRRLVDRCAGRDVLEAGCGEG-YGAALIAEVARSVIGLDYDESAVAHV
MAB_3364|M.abscessus_ATCC_1997 VVYERLSSLCAGRRVLEAGSGEG-YGADMLAQVAQHVIGLDYDESAIAHV
Mb3056|M.bovis_AF2122/97 VVYQRLAPRCTARDVLEAGCGEG-YGADLIACVARQVIAVDYDETAVAHV
Rv3030|M.tuberculosis_H37Rv VVYQRLAPRCTARDVLEAGCGEG-YGADLIACVARQVIAVDYDETAVAHV
MLBr_01713|M.leprae_Br4923 VVYQWLAQRCTGCEVLEAGCGEG-YGADLIVDVAYRVIAVDYDEATVAHV
MUL_1921|M.ulcerans_Agy99 VVYERLAPRCEGRDVLEAGCGEG-YGADMIADVARRVVAVDYDEAAVAHV
MAV_3877|M.avium_104 VVYQRLARHCAGRDVLEAGCGEG-YGADLIAGVARRVIAVDYDEAAVAHV
MMAR_2339|M.marinum_M PEMLRFLP-ADAQCVLELGCGEGVFGAIVKQSTGAEVWGLEYNEEAAQRA
: . *** *.*** :** : .. * .::*: : :.
MSMEG_2350|M.smegmatis_MC2_155 RARYPRVDIRHGNLAELPLPDASVDVVVNFQVIEHLWDQAQFVSECFRVL
TH_2193|M.thermoresistible__bu RARYPRVDIRHGNLVDLPLADASVDVVVNFQVIEHLWDQEKFIAECLRVL
Mflv_4260|M.gilvum_PYR-GCK RARYPRVDMRHGNLAELPLESGSVDVVVNFQVIEHLWDQGQFVAECARVL
Mvan_2098|M.vanbaalenii_PYR-1 RARYPRVDMRHGNLAALPLPTASVDVVVNFQVIEHLWDQGQFVAECARVL
MAB_3364|M.abscessus_ATCC_1997 KARYPRVDMRAGNLADLPLADGSVDVVVNFQVIEHLWDQGQFIRECARVL
Mb3056|M.bovis_AF2122/97 RSRYPRVEVMQANLAELPLPDASVDVVVNFQVIEHLWDQARFVRECARVL
Rv3030|M.tuberculosis_H37Rv RSRYPRVEVMQANLAELPLPDASVDVVVNFQVIEHLWDQARFVRECARVL
MLBr_01713|M.leprae_Br4923 RSRYPRVQVMQANLIELPLADASVDVVVNFQVIEHLWNQAQFVSECARVL
MUL_1921|M.ulcerans_Agy99 RHRYPRVEVMQANLADLPLPDASMDVVVNFQVIEHLWDQGQFVRECARVL
MAV_3877|M.avium_104 RGRYPRVDVMQANLAQLPLPDSSVDVVVNFQVIEHLWDQTQFVVECARVL
MMAR_2339|M.marinum_M GELLDHVLAGDASTRIAELPDNYFDAVVCNDVLEHLVDPLDTLKRLRPKL
:* .. * .*.** :*:*** : : . *
MSMEG_2350|M.smegmatis_MC2_155 RPGGVFLVSTPNRITFSPGRDTPLNPFHTRELNAAELTELLETAGFEVED
TH_2193|M.thermoresistible__bu RPGGQLLVSTPNRITFTPGSDTPVNPFHTRELNAAELTELLTDGGFGLSA
Mflv_4260|M.gilvum_PYR-GCK RPGGVLLMSTPNRITFSPGLDAPVNPFHTRELDAAELTELIESAGFAMET
Mvan_2098|M.vanbaalenii_PYR-1 RPGGVLLMSTPNRITFSPGRDTPVNPFHTRELNAAELTELIESADFAMEA
MAB_3364|M.abscessus_ATCC_1997 RPGGSLLISTPNRITFSPGRDTPLNPFHTRELNGAELTELLEENGFSLES
Mb3056|M.bovis_AF2122/97 RGSGLLMVSTPNRITFSPGRDTPINPFHTRELNADELTSLLIDAGFVDVA
Rv3030|M.tuberculosis_H37Rv RGSGLLMVSTPNRITFSPGRDTPINPFHTRELNADELTSLLIDAGFVDVA
MLBr_01713|M.leprae_Br4923 RPSGLLMVSTPNRITFSPGRDTSINPFHTRELNADELTQLLITADFSEVA
MUL_1921|M.ulcerans_Agy99 RPSGVLMVSTPNRITFSPGRDTPINPFHTRELNAAELTELLLEAGFAEVT
MAV_3877|M.avium_104 RPSGLLLMSTPNRITFSPGRDTPINPFHTRELNAVELTELLVGGGFRDVS
MMAR_2339|M.marinum_M KPDGVVVASIPN-IRYLPALG---KVVFRRDFPQEDF-------GIFDRT
: .* .: * ** * : *. . : .. *:: :: .:
MSMEG_2350|M.smegmatis_MC2_155 TLGVFHGAGLAELDARHGGSIIEAQVQRAVADAPWDEQLLADVAAVRTDD
TH_2193|M.thermoresistible__bu MYGVFHGPRLAELDARHGGSIIDAQISRALADEPWPAELLADVASVRTED
Mflv_4260|M.gilvum_PYR-GCK MLGVFHGPPLVELDARHGGSIIEAQIARAVADAPWPPDLLADVESVTIDH
Mvan_2098|M.vanbaalenii_PYR-1 MLGVFHGRRLAEIDARHGGSVIDAQIARALADAPWPAELLADIESVTIED
MAB_3364|M.abscessus_ATCC_1997 MNGVFHGPRLRELDAKYGGSIIDAQIERAVAGAPWPQELLADITGLTTED
Mb3056|M.bovis_AF2122/97 MCGLFHGPRLRDMDARHGGSIIDAQIMRAVAGAPWPPELAADVAAVTTAD
Rv3030|M.tuberculosis_H37Rv MCGLFHGPRLRDMDARHGGSIIDAQIMRAVAGAPWPPELAADVAAVTTAD
MLBr_01713|M.leprae_Br4923 MYGLFHGPRLKEMDARHGGSIIDAQIARAVSNAPWPPELVADVAAVTAAD
MUL_1921|M.ulcerans_Agy99 MYGLFHGPQLRAIDERHGGSIIDAQIARAVADQPWSAELTADVAAVTTAD
MAV_3877|M.avium_104 ISGLFHGPRLREMDARHGGSIIDAQIARAVADAPWPPQLAADVAAVTVDD
MMAR_2339|M.marinum_M HLRFFTRSSMVRMFEDAG-----FSMQRIKGINPFLGPFGALFVALSLGH
.* : : * .: * . *: : * . .: .
MSMEG_2350|M.smegmatis_MC2_155 FDLTPAAERD-------IDDSLDLVAIAVRP-
TH_2193|M.thermoresistible__bu FDLIRCDEDDRDRDGHNIDDSLDLVAIAVRP-
Mflv_4260|M.gilvum_PYR-GCK FDLTAGSDTTP---GRHIDDSLDLVAIAVRP-
Mvan_2098|M.vanbaalenii_PYR-1 FDLVPADGTH-----RDIDDSLDLVAIAVRT-
MAB_3364|M.abscessus_ATCC_1997 FEIVDAGERD-------IDASLDLVAIAVIP-
Mb3056|M.bovis_AF2122/97 FEMVAAG------HDRDIDDSLDLIAIAVRP-
Rv3030|M.tuberculosis_H37Rv FEMVAAG------HDRDIDDSLDLIAIAVRP-
MLBr_01713|M.leprae_Br4923 FDIIEASAGPEASDGRDVDESLDLIAIAVRR-
MUL_1921|M.ulcerans_Agy99 FDIIGAG----LEPAHEIDDSLDLIAIAVRH-
MAV_3877|M.avium_104 FELVPSG--ADAASGHHIDDSLDLIAIAVAP-
MMAR_2339|M.marinum_M FAD---------------GVFLQYACVATPAS
* . *: .:*.