For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSKAPFLSRAAFARVTNPLARGLLRIGLTPDAVTIIGTTASVAGALVLFPMGKLFPGACVVWFFVLFDML DGAMARERGGGTRFGAVLDAACDRISDGAVFGGLLWWVAFGMRDRLLVVATLICLVTSQVISYIKARAEA SGLRGDGGIIERPERLIIVLAGAGVSDFPFIAWPPALPVAMWLLAVTSVITCGQRLYTVWTSPGATDLLV PSAPVRDDDAQGHPRSGDPGKTQR
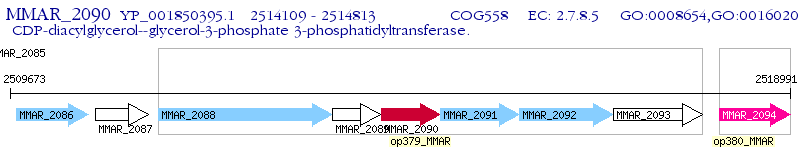
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2090 | pgsA1 | - | 100% (234) | pi synthase PgsA1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2644c | pgsA1 | 1e-104 | 87.92% (207) | CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| M. gilvum PYR-GCK | Mflv_3836 | - | 4e-78 | 69.86% (209) | CDP-alcohol phosphatidyltransferase |
| M. tuberculosis H37Rv | Rv2612c | pgsA1 | 1e-104 | 87.92% (207) | CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| M. leprae Br4923 | MLBr_00454 | pgsA | 1e-92 | 72.17% (230) | putative phosphatidyltransferase |
| M. abscessus ATCC 19977 | MAB_2896c | - | 6e-76 | 66.51% (215) | phosphatidylinositol synthase PgsA |
| M. avium 104 | MAV_3488 | - | 2e-91 | 78.74% (207) | CDP-alcohol phosphatidyltransferase |
| M. smegmatis MC2 155 | MSMEG_2933 | - | 2e-77 | 67.92% (212) | phosphatidylinositol synthase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3254 | pgsA1 | 1e-132 | 99.15% (234) | pi synthase PgsA1 |
| M. vanbaalenii PYR-1 | Mvan_2560 | - | 2e-76 | 68.27% (208) | CDP-alcohol phosphatidyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3836|M.gilvum_PYR-GCK MSNFYLMTRAAYAKLSRPVAKGALKVGLTPDTVTILGTAGSVLAALTLFP
Mvan_2560|M.vanbaalenii_PYR-1 MSDFYLMTRAAYVKLSRPVARGALKLGLSPDSVTILGTAGSVLGALTLFP
MSMEG_2933|M.smegmatis_MC2_155 MSNVYLMTRAAYVKLSRPVAKAALRAGLTPDIVTLAGTAAAVIGALTLFP
MMAR_2090|M.marinum_M MSKAPFLSRAAFARVTNPLARGLLRIGLTPDAVTIIGTTASVAGALVLFP
MUL_3254|M.ulcerans_Agy99 MSKAPFLSRAAFARVTNPLARGLLRIGLTPDAVTIIGTTASVAGALVLFP
Mb2644c|M.bovis_AF2122/97 MSKLPFLSRAAFARITTPIARGLLRVGLTPDVVTILGTTASVAGALTLFP
Rv2612c|M.tuberculosis_H37Rv MSKLPFLSRAAFARITTPIARGLLRVGLTPDVVTILGTTASVAGALTLFP
MLBr_00454|M.leprae_Br4923 MSKVPFLSRATFTRLTTPTARALLRVGLTPDAITVLGTIVAASGALVLFP
MAV_3488|M.avium_104 MSKVPFLSRAAFARLTTPTARACLRLGLTPDVVTVAGTIVAVAGALILFP
MAB_2896c|M.abscessus_ATCC_199 MSG--LLSRETFAKITNPLASALLRAGFTPDTVTIFGTAASVVAALTLFP
** :::* ::.::: * * . *: *::** :*: ** :. .** ***
Mflv_3836|M.gilvum_PYR-GCK IGQLWWGAVAVSFFVLADMLDGAMARERGGGTRFGAVLDAACDRISDGAV
Mvan_2560|M.vanbaalenii_PYR-1 IGQLFAGAWVVAFFVLADMLDGAMARQSGGGTRFGAVLDATCDRISDGAV
MSMEG_2933|M.smegmatis_MC2_155 IGQLWWGAVVVSFFVLADMLDGAMAREQGGGTRFGAVLDATCDRLGDGAV
MMAR_2090|M.marinum_M MGKLFPGACVVWFFVLFDMLDGAMARERGGGTRFGAVLDAACDRISDGAV
MUL_3254|M.ulcerans_Agy99 MGKLFPGACVVWFFVLFDMLDGAMARERGGGTRFGAVLDAACDRISDGAV
Mb2644c|M.bovis_AF2122/97 MGKLFAGACVVWFFVLFDMLDGAMARERGGGTRFGAVLDATCDRISDGAV
Rv2612c|M.tuberculosis_H37Rv MGKLFAGACVVWFFVLFDMLDGAMARERGGGTRFGAVLDATCDRISDGAV
MLBr_00454|M.leprae_Br4923 IGKLFAGTLVVWFFVLFDMLDGAMARERGGGTRYGAVLDATCDRISDGAV
MAV_3488|M.avium_104 IGELFVGTLVVWFFVLFDMLDGAMARERGGGTRFGAVLDATCDRVSDGAV
MAB_2896c|M.abscessus_ATCC_199 TGHLFWGGMAVWLFAMFDMLDGAMARARGGGTRFGAVLDATCDRVADGAV
*.*: * .* :*.: ********* *****:******:***:.****
Mflv_3836|M.gilvum_PYR-GCK FCGLLWWAAFGLQSAELVVATMICLVSSQVISYIKARAEASGLSGDGGLI
Mvan_2560|M.vanbaalenii_PYR-1 FCGLLWWAAFGLHSTGLVVATMICLVSSQVISYIKARAEASGLSGDGGLI
MSMEG_2933|M.smegmatis_MC2_155 FAGLTWWAAFGLDSPSLVVATLICLVTSQVISYIKARAEASGLRGDGGII
MMAR_2090|M.marinum_M FGGLLWWVAFGMRDRLLVVATLICLVTSQVISYIKARAEASGLRGDGGII
MUL_3254|M.ulcerans_Agy99 FSGLLWWVAFGMRDRLLVVATLICLVISQVISYIKARAEASGLRGDGGII
Mb2644c|M.bovis_AF2122/97 FCGLLWWIAFHMRDRPLVIATLICLVTSQVISYIKARAEASGLRGDGGFI
Rv2612c|M.tuberculosis_H37Rv FCGLLWWIAFHMRDRPLVIATLICLVTSQVISYIKARAEASGLRGDGGFI
MLBr_00454|M.leprae_Br4923 FCGLLWWMAFGLHSKHLVVATLICLVTSQVISYIKARAEASGLRGDDGLI
MAV_3488|M.avium_104 FGGLLWWVVFGLHDEPLAAATLICLVTSQVISYIKARAEASGLRGDGGII
MAB_2896c|M.abscessus_ATCC_199 FAGLVWWAAFGWGSTSLVVATLICMITSQVISYVKARAEASGLRADGGLI
* ** ** .* . *. **:**:: ******:********* .*.*:*
Mflv_3836|M.gilvum_PYR-GCK ERPERLVIVLVGAGLAGFPLFPLPWLLHVAMWLLAVTSLITVGQRLHSVR
Mvan_2560|M.vanbaalenii_PYR-1 ERPERLVIILVGACFSDLPFFPLPWLLDVAMWVLALSSVVTVGQRLHSVR
MSMEG_2933|M.smegmatis_MC2_155 ERPERLVIVLIGAGLSDLPFFPLPWTLHVAMWVLAVASVVTLLQRVHAVR
MMAR_2090|M.marinum_M ERPERLIIVLAGAGVSDFPFIAWPPALPVAMWLLAVTSVITCGQRLYTVW
MUL_3254|M.ulcerans_Agy99 ERPERLIIVLAGAGVSDFPFIAWPPALPVAMWLLAVTSVITCGQRLYTVW
Mb2644c|M.bovis_AF2122/97 ERPERLIIVLTGAGVSDFPFVPWPPALSVGMWLLAVASVITCVQRLHTVW
Rv2612c|M.tuberculosis_H37Rv ERPERLIIVLTGAGVSDFPFVPWPPALSVGMWLLAVASVITCVQRLHTVW
MLBr_00454|M.leprae_Br4923 ERPERLIIVLVGAGLSDFPVYSLPWALSVAMWLLVVASLITCAQRLHTVH
MAV_3488|M.avium_104 ERPERLIIVLVGAGLSDLPGYPLPWALPVAMWALAAASLVTCAQRLHTVR
MAB_2896c|M.abscessus_ATCC_199 ERPERLIIVLAGAIFSGG--FGVQWPLHTAMWVLAVASLVTVAQRMHAVR
******:*:* ** .:. * ..** *. :*::* **:::*
Mflv_3836|M.gilvum_PYR-GCK TSAAAMEPLSVPD--------------KP-----ETTEQ
Mvan_2560|M.vanbaalenii_PYR-1 TSAAAMEPLKVPEA-----------GEKP-----ETTEQ
MSMEG_2933|M.smegmatis_MC2_155 TSPGAMEPLHPAN------------GEKP-----ETSEP
MMAR_2090|M.marinum_M TSPGATDLLVPSAPVRDDDAQGHPRSGDP-----GKTQR
MUL_3254|M.ulcerans_Agy99 TSPGATDLLVPSAPVRDDDAQGHPRSGDP-----GKTQR
Mb2644c|M.bovis_AF2122/97 TSPGAID-----------------RMAIP-----GKGDR
Rv2612c|M.tuberculosis_H37Rv TSPGAID-----------------RMAIP-----GKGDR
MLBr_00454|M.leprae_Br4923 TSPGATDRIARPGYPSASDRTNATNGGQPGPGNLGTGEP
MAV_3488|M.avium_104 TSPGATDR--------------------------GSDAP
MAB_2896c|M.abscessus_ATCC_199 TSPGALDLLPNSD-----------AGQDT----AETNQP
**..* : .