For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAQADREGEMAQPIARGIVGRVDSDLDAQSPAADLVRVYLNGIGKTALLNAEGEVELAKRIEAGLYAEHL LATRKRLGDNRKRELAAVVRDGAEARRHLLEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFD YTKGFKFSTYATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREATDEELAAESGIP VEKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAMSAENAVIAELLHTDIRSVLATLDEREHQVIR LRFGLDDGQPRTLDQIGKLFGLSRERVRQIERDVMCKLRHGERADRLRSYAS*
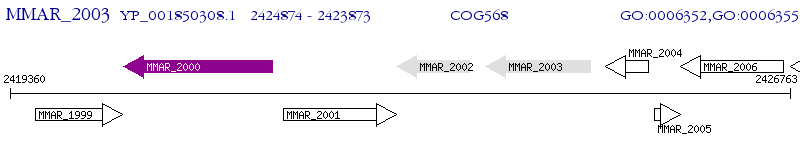
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2003 | sigB | - | 100% (333) | RNA polymerase sigma factor SigB |
| M. marinum M | MMAR_2011 | sigA | e-106 | 63.06% (314) | RNA polymerase sigma factor SigA |
| M. marinum M | MMAR_1248 | sigF | 8e-19 | 30.86% (243) | alternate RNA polymerase sigma factor SigF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2729 | sigB | 1e-170 | 94.43% (323) | RNA polymerase sigma factor SigB |
| M. gilvum PYR-GCK | Mflv_3948 | - | 1e-164 | 89.19% (333) | RNA polymerase sigma factor SigB |
| M. tuberculosis H37Rv | Rv2710 | sigB | 1e-170 | 94.43% (323) | RNA polymerase sigma factor SigB |
| M. leprae Br4923 | MLBr_01014 | - | 1e-169 | 94.67% (319) | RNA polymerase sigma factor SigB |
| M. abscessus ATCC 19977 | MAB_3028 | - | 1e-155 | 85.76% (323) | RNA polymerase sigma factor SigB |
| M. avium 104 | MAV_3603 | - | 1e-168 | 94.04% (319) | RNA polymerase sigma factor SigB |
| M. smegmatis MC2 155 | MSMEG_2752 | - | 1e-162 | 91.22% (319) | RNA polymerase sigma factor SigB |
| M. thermoresistible (build 8) | TH_0829 | sigB | 1e-127 | 71.92% (317) | RNA POLYMERASE SIGMA FACTOR SIGB |
| M. ulcerans Agy99 | MUL_3350 | sigB | 1e-180 | 99.38% (323) | RNA polymerase sigma factor SigB |
| M. vanbaalenii PYR-1 | Mvan_2451 | - | 1e-164 | 89.49% (333) | RNA polymerase sigma factor SigB |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3948|M.gilvum_PYR-GCK ----MNVPPDQEAAMANATTSRFDQDLDAQSPAADLVRVYLNGIGKTALL
Mvan_2451|M.vanbaalenii_PYR-1 ----MKVPPDQEAAMANATTSRIDQDLDAQSPAADLVRVYLNGIGKTALL
MSMEG_2752|M.smegmatis_MC2_155 --------------MANATTSRVDTDLDAQSPAADLVRVYLNGIGKTALL
Mb2729|M.bovis_AF2122/97 ----------MADAPTRATTSRVDSDLDAQSPAADLVRVYLNGIGKTALL
Rv2710|M.tuberculosis_H37Rv ----------MADAPTRATTSRVDSDLDAQSPAADLVRVYLNGIGKTALL
MAV_3603|M.avium_104 ----------MAN----ASTSRFDGDLDAQSPAADLVRVYLNGIGKTALL
MMAR_2003|M.marinum_M MTAQADREGEMAQPIARGIVGRVDSDLDAQSPAADLVRVYLNGIGKTALL
MUL_3350|M.ulcerans_Agy99 ----------MAQPIARGIVGRVDSDLDAQSPAADLVRVYLNGIGKTALL
MLBr_01014|M.leprae_Br4923 --------------MAEATTNRVDSDLDAQSPAADLVRVYLNGIGKTALL
MAB_3028|M.abscessus_ATCC_1997 ----------MANATTRPARTTESEDLDAQSPAADLVRVYLNGIGKTALL
TH_0829|M.thermoresistible__bu --------------MANATTSRVDSDLDAQSPAADLVRVYLNGIGKTALL
. *************************
Mflv_3948|M.gilvum_PYR-GCK TAADEVELAKRIEAGLYAQHLLATRKRLGENRKRDLAAVVRDGEAARRHL
Mvan_2451|M.vanbaalenii_PYR-1 NAADEVELAKRIEAGLYAQHLLDTRKRLGENRKRDLAAVVRDGEAARRHL
MSMEG_2752|M.smegmatis_MC2_155 NAADEVELAKRIEAGLYAQHLLDTKKRLGEARKRDLQAVVRDGEAARRHL
Mb2729|M.bovis_AF2122/97 NAAGEVELAKRIEAGLYAEHLLETRKRLGENRKRDLAAVVRDGEAARRHL
Rv2710|M.tuberculosis_H37Rv NAAGEVELAKRIEAGLYAEHLLETRKRLGENRKRDLAAVVRDGEAARRHL
MAV_3603|M.avium_104 NAAGEVELAKRIEAGLYAEHLLETRKRLGENRKRDLEAVVRDGQAARRHL
MMAR_2003|M.marinum_M NAEGEVELAKRIEAGLYAEHLLATRKRLGDNRKRELAAVVRDGAEARRHL
MUL_3350|M.ulcerans_Agy99 NAEGEVELAKRIEAGLYAEHLLATRKRLGDNRKRELAAVVRDGAEARRHL
MLBr_01014|M.leprae_Br4923 NAADEVELAKRIEAGLYAEHLLQTRKRFGEGRKRALAAVARDGAAARRHL
MAB_3028|M.abscessus_ATCC_1997 TAEDEVELAKRIEAGLYAQHLLDTKKRLGEARKKDLAVIVREGNAARSHL
TH_0829|M.thermoresistible__bu DAAEEVELAKRIEAGLYAQHLLKTKKRLGQNRKRDLEAIVRDGEEARRHL
* **************:*** *:**:*: **: * .:.*:* ** **
Mflv_3948|M.gilvum_PYR-GCK LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYAKGFKFST
Mvan_2451|M.vanbaalenii_PYR-1 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYAKGFKFST
MSMEG_2752|M.smegmatis_MC2_155 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
Mb2729|M.bovis_AF2122/97 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
Rv2710|M.tuberculosis_H37Rv LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
MAV_3603|M.avium_104 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
MMAR_2003|M.marinum_M LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
MUL_3350|M.ulcerans_Agy99 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
MLBr_01014|M.leprae_Br4923 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
MAB_3028|M.abscessus_ATCC_1997 LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYAKGFKFST
TH_0829|M.thermoresistible__bu LEANLRLVVSLAKRYTGRGMPLLDLIQEGNLGLIRAMEKFDYTKGFKFST
******************************************:*******
Mflv_3948|M.gilvum_PYR-GCK YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
Mvan_2451|M.vanbaalenii_PYR-1 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
MSMEG_2752|M.smegmatis_MC2_155 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
Mb2729|M.bovis_AF2122/97 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQHLGREAT
Rv2710|M.tuberculosis_H37Rv YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQHLGREAT
MAV_3603|M.avium_104 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
MMAR_2003|M.marinum_M YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
MUL_3350|M.ulcerans_Agy99 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREIHQNLGREAT
MLBr_01014|M.leprae_Br4923 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKREMHQNLGREAT
MAB_3028|M.abscessus_ATCC_1997 YATWWIRQAITRGMADQSRTIRLPVHLVEQVNKLARIKRELHQRLGREAS
TH_0829|M.thermoresistible__bu YATWWIRQAITRAMADQARTIRIPVHMVEVINKLGRIQRELLQDLGREPT
************.****:****:***:** :***.**:**: * ****.:
Mflv_3948|M.gilvum_PYR-GCK DEELAEESGIPVEKINDLLEHSRDPVSLDMPVGSDEEAPLGDFIEDAEAM
Mvan_2451|M.vanbaalenii_PYR-1 DEELAAESGIPVEKITDLLEHSRDPVSLDMPVGSDEEAPLGDFIEDAEAM
MSMEG_2752|M.smegmatis_MC2_155 DEELAEESGIPVEKINDLLEHSRDPVSLDMPVGTDEEAPLGDFIEDSEAM
Mb2729|M.bovis_AF2122/97 DEELAAESGIPIDKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAM
Rv2710|M.tuberculosis_H37Rv DEELAAESGIPIDKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAM
MAV_3603|M.avium_104 DEELAAESGIPIDKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAM
MMAR_2003|M.marinum_M DEELAAESGIPVEKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAM
MUL_3350|M.ulcerans_Agy99 DEELAAESGIPVEKINDLLEHSRDPVSLDMPVGSDEEAPLGDFIEDAEAM
MLBr_01014|M.leprae_Br4923 DEELAAESGIPIEKINDLLEHSRDPVSLDMPVGSEEEAPLGDFIEDAEAM
MAB_3028|M.abscessus_ATCC_1997 DEELAEESGIPVEKIGDLLDHSRDPVSLDMPVGTDEEAPLGDFIEDTEAM
TH_0829|M.thermoresistible__bu PEELAKEMDITPEKVLEIQQYAREPISLDQTIGDEGDSQLGDFIEDSEAV
**** * .*. :*: :: :::*:*:*** .:* : :: *******:**:
Mflv_3948|M.gilvum_PYR-GCK SAENAVISELLHTDIRYVLATLDEREQQVIRLRFGLDDGQPRTLDQIGKL
Mvan_2451|M.vanbaalenii_PYR-1 SAENAVISELLHTDIRYVLATLDEREQQVIRLRFGLDDGQPRTLDQIGKL
MSMEG_2752|M.smegmatis_MC2_155 SAENAVISELLHTDIRYVLATLDEREQQVIRLRFGLDDGQPRTLDQIGKL
Mb2729|M.bovis_AF2122/97 SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
Rv2710|M.tuberculosis_H37Rv SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
MAV_3603|M.avium_104 SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
MMAR_2003|M.marinum_M SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
MUL_3350|M.ulcerans_Agy99 SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
MLBr_01014|M.leprae_Br4923 SAENAVIAELLHTDIRSVLATLDEREHQVIRLRFGLDDGQPRTLDQIGKL
MAB_3028|M.abscessus_ATCC_1997 SAENAVIAELLHSDVRSVLATLDEREQQVIRLRYGLDDGQARTLDQIGKL
TH_0829|M.thermoresistible__bu VAVDAVSFTLLQDQLQSVLETLSEREAGVVRLRFGLTDGQPRTLDEIGQV
* :** **: ::: ** **.*** *:***:** ***.****:**::
Mflv_3948|M.gilvum_PYR-GCK FGLSRERVRQIEREVMAKLRNGDRADRLRSYAS
Mvan_2451|M.vanbaalenii_PYR-1 FGLSRERVRQIEREVMAKLRNGERADRLRSYAS
MSMEG_2752|M.smegmatis_MC2_155 FGLSRERVRQIEREVMAKLRNGERADRLRSYAS
Mb2729|M.bovis_AF2122/97 FGLSRERVRQIERDVMSKLRHGERADRLRSYAS
Rv2710|M.tuberculosis_H37Rv FGLSRERVRQIERDVMSKLRHGERADRLRSYAS
MAV_3603|M.avium_104 FGLSRERVRQIERDVMSKLRNGERADRLRSYAS
MMAR_2003|M.marinum_M FGLSRERVRQIERDVMCKLRHGERADRLRSYAS
MUL_3350|M.ulcerans_Agy99 FGLSRERVRQIERDVMCKLRHGERADRLRSYAS
MLBr_01014|M.leprae_Br4923 FGLSRERVRQIERDVMCKLRNGERADRLRSYAS
MAB_3028|M.abscessus_ATCC_1997 FGLSRERVRQIEREVMAKLRHGDRAERLRSYAS
TH_0829|M.thermoresistible__bu YGVTRERIRQIESKTMSKLRHPSRSQVLRDYLD
:*::***:**** ..*.***: .*:: **.* .