For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTILLVNPPGARRNGRSYLSEQKLGDPWVYSTMPMEHLGMMSIKAYAKTQGIEIATVNGLVAGHSSVQET WSAIENAVRCSGKPRLVGFSCIDTLPEVLWLAQRVRETWDDVQIALGNAIATLNYERILRQHDCFDFVIV GDAEVGFTKLANAVANGAALDDVPGLARRDDRGQIICSPSRLIDLDELPRPARDELPSVLADGFSAAVFS TRGCPYRCTFCGTGAMSAMLGRNSYRAKSVDAVVDEIAYLKSDFDIGFLCITDDLFISKHPSSQQRAAEF ADAMINSGVDVKFMMDIRLDSVVDLELFKHLHKAGLRRVFVGLETGSYDQLRAYRKQIINRGQDAADTIN ALQQVGVDVIPGTIMFHPTVQPDELRETARLLRATKFISTYKFMGKITPYPGTPLYQEYSDLGYLREEWP LGEWDFVDPEAARVYADVCSLIAPRPDLPYDEAEKFFLARLDEWDDVIAARNEQTAADGTGTLEPPGGPQ TPELLSLT
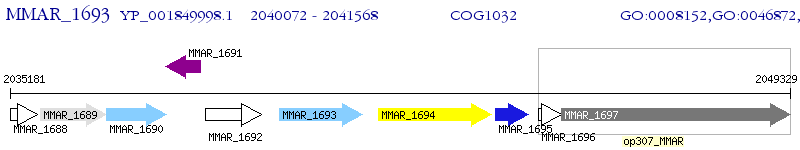
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1693 | - | - | 100% (498) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0219c | - | 0.0 | 76.33% (431) | methyltransferase (methylase) |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0213c | - | 0.0 | 76.33% (431) | methyltransferase (methylase) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0219c|M.bovis_AF2122/97 ----------------------------------------MSIKAYAKTQ
Rv0213c|M.tuberculosis_H37Rv ----------------------------------------MSIKAYAKTQ
MMAR_1693|M.marinum_M MTILLVNPPGARRNGRSYLSEQKLGDPWVYSTMPMEHLGMMSIKAYAKTQ
**********
Mb0219c|M.bovis_AF2122/97 GIAVTSVNGLVAGHGSVQETWLAMQSAAALSGTPRLVGFSCIDTFPEVLW
Rv0213c|M.tuberculosis_H37Rv GIAVTSVNGLVAGHGSVQETWLAMQSAAALSGTPRLVGFSCIDTFPEVLW
MMAR_1693|M.marinum_M GIEIATVNGLVAGHSSVQETWSAIENAVRCSGKPRLVGFSCIDTLPEVLW
** :::********.****** *::.*. **.***********:*****
Mb0219c|M.bovis_AF2122/97 LAQRARQAWDGVRIVIGNAMATLNYERILRQHDCFDYVVVGDGEVAFTKL
Rv0213c|M.tuberculosis_H37Rv LAQRARQAWDGVRIVIGNAMATLNYERILRQHDCFDYVVVGDGEVAFTKL
MMAR_1693|M.marinum_M LAQRVRETWDDVQIALGNAIATLNYERILRQHDCFDFVIVGDAEVGFTKL
****.*::**.*:*.:***:****************:*:***.**.****
Mb0219c|M.bovis_AF2122/97 ALALANDAAVDDVPGLARRSEQGQILRTPSSLVDLDELPRPARDELPTVL
Rv0213c|M.tuberculosis_H37Rv ALALANDAAVDDVPGLARRSEQGQILRTPSSLVDLDELPRPARDELPTVL
MMAR_1693|M.marinum_M ANAVANGAALDDVPGLARRDDRGQIICSPSRLIDLDELPRPARDELPSVL
* *:**.**:*********.::***: :** *:**************:**
Mb0219c|M.bovis_AF2122/97 ADGFAASVFSTRGCPYRCTFCGTGAMSAMLGKDSYRAKSVDAVVDEIDYL
Rv0213c|M.tuberculosis_H37Rv ADGFAASVFSTRGCPYRCTFCGTGAMSAMLGKDSYRAKSVDAVVDEIDYL
MMAR_1693|M.marinum_M ADGFSAAVFSTRGCPYRCTFCGTGAMSAMLGRNSYRAKSVDAVVDEIAYL
****:*:************************::************** **
Mb0219c|M.bovis_AF2122/97 VSDYDVNFLSITDDLFISKHPGSQQRAADFANAVLRRGISVNFMVDIRLD
Rv0213c|M.tuberculosis_H37Rv VSDYDVNFLSITDDLFISKHPGSQQRAADFANAVLRRGISVNFMVDIRLD
MMAR_1693|M.marinum_M KSDFDIGFLCITDDLFISKHPSSQQRAAEFADAMINSGVDVKFMMDIRLD
**:*:.**.***********.******:**:*::. *:.*:**:*****
Mb0219c|M.bovis_AF2122/97 SVVDLDLFKHLHRAGLRRVFIGVETGSYEQLRAYRKQILTRGQDAADTIN
Rv0213c|M.tuberculosis_H37Rv SVVDLDLFKHLHRAGLRRVFIGVETGSYEQLRAYRKQILTRGQDAADTIN
MMAR_1693|M.marinum_M SVVDLELFKHLHKAGLRRVFVGLETGSYDQLRAYRKQIINRGQDAADTIN
*****:******:*******:*:*****:*********:.**********
Mb0219c|M.bovis_AF2122/97 ALQQLGIDVIPGTIMFHPTVQPDELRETVRLLRATKYTVGFKFMSRIVPY
Rv0213c|M.tuberculosis_H37Rv ALQQLGIDVIPGTIMFHPTVQPDELRETVRLLRATKYTVGFKFMSRIVPY
MMAR_1693|M.marinum_M ALQQVGVDVIPGTIMFHPTVQPDELRETARLLRATKFISTYKFMGKITPY
****:*:*********************.*******: :***.:*.**
Mb0219c|M.bovis_AF2122/97 PGTPLYQAYSDAGYLTAKWPLGQWEFVDPEASRVYADVVAKVAPDVGISF
Rv0213c|M.tuberculosis_H37Rv PGTPLYQAYSDAGYLTAKWPLGQWEFVDPEASRVYADVVAKVAPDVGISF
MMAR_1693|M.marinum_M PGTPLYQEYSDLGYLREEWPLGEWDFVDPEAARVYADVCSLIAPRPDLPY
******* *** *** :****:*:******:****** : :** .:.:
Mb0219c|M.bovis_AF2122/97 DEAEAYFLSRLDEWENVIAGRIAEATS---------------------
Rv0213c|M.tuberculosis_H37Rv DEAEAYFLSRLDEWENVIAGRIAEATS---------------------
MMAR_1693|M.marinum_M DEAEKFFLARLDEWDDVIAARNEQTAADGTGTLEPPGGPQTPELLSLT
**** :**:*****::***.* ::::