For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVAKAVALMVLAVGLAAPAAADSTDDAFLTNLNTSGMDFGSPDKAIQVAKTIVCGSLHDNPNASNADLIA KVTKATDWPALDAAYFTGAAIQAYCPQYGSLTPPSLPSKVPTAPSAPPAPPSTSVVQTA
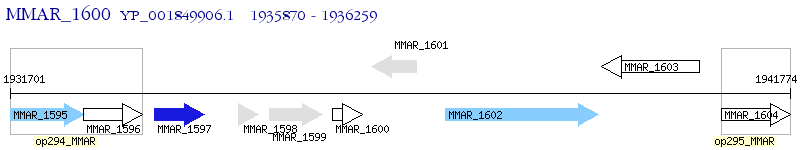
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1600 | - | - | 100% (129) | hypothetical protein MMAR_1600 |
| M. marinum M | MMAR_1178 | - | 3e-12 | 33.61% (122) | hypothetical protein MMAR_1178 |
| M. marinum M | MMAR_1233 | - | 2e-10 | 36.63% (101) | hypothetical protein MMAR_1233 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1302c | - | 2e-10 | 35.71% (98) | hypothetical protein Mb1302c |
| M. gilvum PYR-GCK | Mflv_2012 | - | 2e-09 | 34.88% (86) | hypothetical protein Mflv_2012 |
| M. tuberculosis H37Rv | Rv1271c | - | 2e-10 | 35.71% (98) | hypothetical protein Rv1271c |
| M. leprae Br4923 | MLBr_00676 | - | 3e-08 | 29.66% (118) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3549 | - | 6e-15 | 41.28% (109) | hypothetical protein MAV_3549 |
| M. smegmatis MC2 155 | MSMEG_4689 | - | 7e-08 | 34.82% (112) | hypothetical protein MSMEG_4689 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1430 | - | 4e-12 | 33.61% (122) | hypothetical protein MUL_1430 |
| M. vanbaalenii PYR-1 | Mvan_4716 | - | 6e-12 | 36.46% (96) | hypothetical protein Mvan_4716 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1600|M.marinum_M -----------------------------------------MVAKAVALM
MAV_3549|M.avium_104 ------------------------------MTHTKAGRAAWPAACAVVLS
Mflv_2012|M.gilvum_PYR-GCK ------------------------------------MRGTFFITT-LAAA
Mvan_4716|M.vanbaalenii_PYR-1 ------------------------------------MRGKLIVSAAVAAA
Mb1302c|M.bovis_AF2122/97 ----------------------------MLSPLSPRIIAAFTTAVGAAAI
Rv1271c|M.tuberculosis_H37Rv ----------------------------MLSPLSPRIIAAFTTAVGAAAI
MLBr_00676|M.leprae_Br4923 MFLRLMLSALKALQRLGAVMNSLARIDHWIWLFRCQPLTIRLLVATAALF
MUL_1430|M.ulcerans_Agy99 -----------------------------MRAWRYQPLTVRLLAGAAGLL
MSMEG_4689|M.smegmatis_MC2_155 ----------------------------MSARLPAVLVTAAALAAGVGAT
MMAR_1600|M.marinum_M VLAVGLAAP-AAADSTDDAFLTNLNTSGMDFGSPDKAIQVAKTIVCGSLH
MAV_3549|M.avium_104 AAALLCAAP-AAADEADDAFLAGLAKGGITMFDDDDAIAMGHS-VCSSID
Mflv_2012|M.gilvum_PYR-GCK GLATALAAP-AFADETDDIFIAALEQEGIPFSTAGNAIELAGA-VCEYAA
Mvan_4716|M.vanbaalenii_PYR-1 GLATALAAP-AFADETDDIFISALQNQGIPFSTAENAIELASA-VCDYAA
Mb1302c|M.bovis_AF2122/97 GLAVATAGT-AGANTKDEAFIAQMESIGVTFSSPQVATQQAQL-VCKKLA
Rv1271c|M.tuberculosis_H37Rv GLAVATAGT-AGANTKDEAFIAQMESIGVTFSSPQVATQQAQL-VCKKLA
MLBr_00676|M.leprae_Br4923 TAATAFEVP-AEADAIDDTFIKALNHAGVNFGEPRSAMTMGHY-VCPILA
MUL_1430|M.ulcerans_Agy99 TASAALAAP-AEADSVDDAFINALNAAGVNYGDAGGAEALGRS-VCPVLS
MSMEG_4689|M.smegmatis_MC2_155 PAHADPAAPGAPGNPVDRSFLTALNNAGVGYADPDAAVRLGQD-VCPMLV
. * .: * *: : *: * . **
MMAR_1600|M.marinum_M DNPNASNADLIAKVTKATDWPALDAAYFTGAAIQAYCPQYGSLTPP-SLP
MAV_3549|M.avium_104 ANPNVS--MLALRLTKQTPLTPKQSGYFIGLSVASYCPQYKDDVDP-SLG
Mflv_2012|M.gilvum_PYR-GCK AGQDKT--QIALEIMEPAGWSPEQSGFFVGAATQSYCP------------
Mvan_4716|M.vanbaalenii_PYR-1 AGQDPT--QIALEIMEPAGWTAEQSGFFVGAATQSYCPS-----------
Mb1302c|M.bovis_AF2122/97 SGETGT--EIAEEVLSQTNLTTKQAAYFVVDATKAYCPQYASQLT-----
Rv1271c|M.tuberculosis_H37Rv SGETGT--EIAEEVLSQTNLTTKQAAYFVVDATKAYCPQYASQLT-----
MLBr_00676|M.leprae_Br4923 KSGGNF-AAAVQRIRGNSDMSPQMAETFAKIAISIYCPTMMANVASGNLP
MUL_1430|M.ulcerans_Agy99 EPGGSFNSAASKVVAGGSGMSPEMAATFTSIAISMYCPSVMANVTSGNLP
MSMEG_4689|M.smegmatis_MC2_155 EPGKNFASVVTKLRGDRSVVSPDVAAFFAGIAISMYCPQMISGIGDGTLL
: .. : * : ***
MMAR_1600|M.marinum_M SKVPTAPSAPPAPPSTSVVQTA
MAV_3549|M.avium_104 WLIP--------PPLM------
Mflv_2012|M.gilvum_PYR-GCK ----------------------
Mvan_4716|M.vanbaalenii_PYR-1 ----------------------
Mb1302c|M.bovis_AF2122/97 ----------------------
Rv1271c|M.tuberculosis_H37Rv ----------------------
MLBr_00676|M.leprae_Br4923 -----SLPPGPGIPGI------
MUL_1430|M.ulcerans_Agy99 LQGIPGVPAIPGVPGVPGF---
MSMEG_4689|M.smegmatis_MC2_155 R-----MLAVPGLGALGR----