For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDTETVLVAGGQDWNEIVEAARGADPGERIVVNMGPQHPSTHGVLRLILEIEGETVTEARCGIGYLHTGI EKNLEYRYWTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDEIPERVNVIRVMMMELNRISSHLVALA TGGMELGAMTPMFVGFRAREIILSLFESITGLRMNSAYIRPGGVAQDLPPDGPTEIAKAIADLRQPLREM GDLLNENAIWKARTQDVGYLDLAGCMALGITGPILRSTGLPHDLRKSEPYCGYENYEFDVITADGCDAYG RYMIRVKEMWESIKIVEQCLDRLRPGPTMIEDRKLAWPADLKVGPDGMGNSPEHIAKIMGSSMEALIHHF KLVTEGIRVPAGQVYVAVESPRGELGVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMSEGGMVADLITAVA SIDPVMGGVDR*
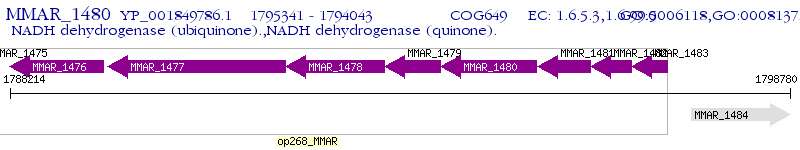
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1480 | nuoD | - | 100% (432) | NADH dehydrogenase I (chain D) NuoD (NADH- ubiquinone oxidoreductase chain D) |
| M. marinum M | MMAR_1659 | hycE | 1e-18 | 26.22% (267) | formate hydrogenase HycE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3172 | nuoD | 0.0 | 92.29% (428) | NADH dehydrogenase subunit D |
| M. gilvum PYR-GCK | Mflv_4484 | - | 0.0 | 80.36% (443) | NADH dehydrogenase subunit D |
| M. tuberculosis H37Rv | Rv3148 | nuoD | 0.0 | 92.29% (428) | NADH dehydrogenase subunit D |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2137 | - | 0.0 | 80.61% (428) | NADH dehydrogenase subunit D |
| M. avium 104 | MAV_4036 | nuoD | 0.0 | 92.06% (428) | NADH dehydrogenase subunit D |
| M. smegmatis MC2 155 | MSMEG_2060 | nuoD | 0.0 | 82.83% (431) | NADH dehydrogenase subunit D |
| M. thermoresistible (build 8) | TH_0634 | nuoD | 0.0 | 81.80% (434) | PROBABLE NADH DEHYDROGENASE I (CHAIN D) NUOD |
| M. ulcerans Agy99 | MUL_2462 | nuoD | 0.0 | 99.54% (432) | NADH dehydrogenase subunit D |
| M. vanbaalenii PYR-1 | Mvan_1882 | - | 0.0 | 81.34% (434) | NADH dehydrogenase subunit D |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2060|M.smegmatis_MC2_155 MSTSTVPPD--GGEKVVVVGGNDWHEVVAAARAG---------AAAQAGE
TH_0634|M.thermoresistible__bu --MSTRP------ETVITLGGQDWDQLVAEARSN---------AADQTGE
Mflv_4484|M.gilvum_PYR-GCK MTTSAQRP-----ERVIVVGGQDWDQVVTAARQMSNSSSRAGDAAEHAGE
Mvan_1882|M.vanbaalenii_PYR-1 MTTSQQPP-----ERVVVVGGQDWDQVVAAARQN---------AAEHAGE
MAB_2137|M.abscessus_ATCC_1997 --MTAQP------EIHLMAGGQDWDEIVTAAGQA-------------TDE
MMAR_1480|M.marinum_M MTDT---------ETVLVAGGQDWNEIVEAARGAD------------PGE
MUL_2462|M.ulcerans_Agy99 MTDT---------ETVLVAGGQDWNEIVEAARGAD------------PGE
Mb3172|M.bovis_AF2122/97 MTAIADSAG-GAGETVLVAGGQDWQQVVDAARSAD------------PGE
Rv3148|M.tuberculosis_H37Rv MTAIADSAG-GAGETVLVAGGQDWQQVVDAARSAD------------PGE
MAV_4036|M.avium_104 MSTTTESTHDGAGETLVVAGGQDWDKVVEAARSAD------------PGE
* : **:**.::* * ..*
MSMEG_2060|M.smegmatis_MC2_155 RIVVNMGPQHPSTHGVLRLILEIEGEIITEARCGIGYLHTGIEKNLEYRN
TH_0634|M.thermoresistible__bu RIVVNMGPQHPSTHGVLRLVLEIEGETVTEARCGVGYLHTGIEKNLEYRT
Mflv_4484|M.gilvum_PYR-GCK RIVVNMGPQHPSTHGVLRLILEIEGETIVEARCGIGYLHTGIEKNLEFRT
Mvan_1882|M.vanbaalenii_PYR-1 RIVVNMGPQHPSTHGVLRLILEIEGEIIVEARCGIGYLHTGIEKNLEFRN
MAB_2137|M.abscessus_ATCC_1997 RIVVNMGPQHPSTHGVLRLILEIEGETVTDVRCGIGYLHTGIEKNLEYRT
MMAR_1480|M.marinum_M RIVVNMGPQHPSTHGVLRLILEIEGETVTEARCGIGYLHTGIEKNLEYRY
MUL_2462|M.ulcerans_Agy99 RIVVNMGPQHPSTHGVLRLILEIEGETVTEARCGIGYLHTGIEKNLEYRY
Mb3172|M.bovis_AF2122/97 RIVVNMGPQHPSTHGVLRLILEIEGETVVEARCGIGYLHTGIEKNLEYRY
Rv3148|M.tuberculosis_H37Rv RIVVNMGPQHPSTHGVLRLILEIEGETVVEARCGIGYLHTGIEKNLEYRY
MAV_4036|M.avium_104 RIVVNMGPQHPSTHGVLRLILEIEGETVTEARCGIGYLHTGIEKNLEYRY
*******************:****** :.:.***:************:*
MSMEG_2060|M.smegmatis_MC2_155 WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDDIPERASVIRVMLME
TH_0634|M.thermoresistible__bu WMQGVTFVTRMDYLSPFFNETVYCLGVEKLLGITEAIPERANVIRVMLME
Mflv_4484|M.gilvum_PYR-GCK WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGVTDDIPERASVIRVMMME
Mvan_1882|M.vanbaalenii_PYR-1 WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDAIPERASVIRVMMME
MAB_2137|M.abscessus_ATCC_1997 WTQGVTFVTRMDYLSPFFNETAYCLGVEQLLGIADEIPERANVIRVLMME
MMAR_1480|M.marinum_M WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDEIPERVNVIRVMMME
MUL_2462|M.ulcerans_Agy99 WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDEIPERVNVIRVMMME
Mb3172|M.bovis_AF2122/97 WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDEIPERVNVIRVLMME
Rv3148|M.tuberculosis_H37Rv WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDEIPERVNVIRVLMME
MAV_4036|M.avium_104 WTQGVTFVTRMDYLSPFFNETAYCLGVEKLLGITDQIPERVNVIRVMMME
* *******************.******:***::: ****..****::**
MSMEG_2060|M.smegmatis_MC2_155 LNRISSHLVALATGGMELGAMSAMFYGFREREEILRVFESITGLRMNHAY
TH_0634|M.thermoresistible__bu LNRISSHLVALATGGMELGAMTAMFLGFREREEILKVFELITGLRMNHAY
Mflv_4484|M.gilvum_PYR-GCK LNRISSHLVALATGGMELGAMTAMFLGFRERELILSVFETITGLRMNSAY
Mvan_1882|M.vanbaalenii_PYR-1 LNRISSHLVALATGGMELGAMTAMFLGFRERELILSVFETITGLRMNNAY
MAB_2137|M.abscessus_ATCC_1997 LNRISSHLVALATGGMELGAMTAMFLGFREREMILKVFEAITGLRMNHAY
MMAR_1480|M.marinum_M LNRISSHLVALATGGMELGAMTPMFVGFRAREIILSLFESITGLRMNSAY
MUL_2462|M.ulcerans_Agy99 LNRISSHLVALAAGGMELGAMTPMFVGFRAREIILSLFESITGLRMNSAY
Mb3172|M.bovis_AF2122/97 LNRISSHLVALATGGMELGAMTPMFVGFRAREIVLTLFEKITGLRMNSAY
Rv3148|M.tuberculosis_H37Rv LNRISSHLVALATGGMELGAMTPMFVGFRAREIVLTLFEKITGLRMNSAY
MAV_4036|M.avium_104 LNRISSHLVALATGGMELGAMTPMFVGFRGREIVLTLFESITGLRMNSAY
************:********:.** *** ** :* :** ******* **
MSMEG_2060|M.smegmatis_MC2_155 IRPGGLAADLPDDAITQVRRLVEILPKRLKDLEDLLNENYIWKARTVGVG
TH_0634|M.thermoresistible__bu IRPGGLAADLPPDAVPRVRELLELLPGRLRDLEDLLSQNHIWRARNEGVG
Mflv_4484|M.gilvum_PYR-GCK IRPGGVAADLPEEGLPQIRELLTLLPTRLRDMENLLNENYIWKARTLGVG
Mvan_1882|M.vanbaalenii_PYR-1 IRPGGVAADLPDEALPQVRDLLTLLPKRLRDMEDLLNENYIWKARTQGIG
MAB_2137|M.abscessus_ATCC_1997 VRPGGLAQDLPDGAEQEVRDLLEILPGRLRDMENLLNENYIWKARTQGVG
MMAR_1480|M.marinum_M IRPGGVAQDLPPDGPTEIAKAIADLRQPLREMGDLLNENAIWKARTQDVG
MUL_2462|M.ulcerans_Agy99 IRPGGVAQDLPPDRPTEIAKAIADLRQPLREMGDLLNENAIWKARTQDVG
Mb3172|M.bovis_AF2122/97 IRPGGVAQDLPPNAATEIAEALKQLRQPLREMGELLNENAIWKARTQGVG
Rv3148|M.tuberculosis_H37Rv IRPGGVAQDLPPNAATEIAEALKQLRQPLREMGELLNENAIWKARTQGVG
MAV_4036|M.avium_104 IRPGGVAQDLPPNAKTEIAQALDQLRQPLREMGDLLNENAIWKARTQDIG
:****:* *** .: : * *::: :**.:* **:**. .:*
MSMEG_2060|M.smegmatis_MC2_155 YLDLTGCMALGITGPILRSTGLPHDLRKAQPYCGYENYEFDVITDDRCDS
TH_0634|M.thermoresistible__bu YLDLTGCMALGVTGPVLRSAGLPHDLRKSQPYCGYENYEFDVITDDRCDA
Mflv_4484|M.gilvum_PYR-GCK YLDLTGCMALGITGPVLRSTGLPHDLRKSQPYCGYQTYDFDVITDDRCDS
Mvan_1882|M.vanbaalenii_PYR-1 YLDLTGCMALGITGPVLRSTGLPHDLRKAQPYCGYETYDFDVVTDDQCDS
MAB_2137|M.abscessus_ATCC_1997 YLDLTGCMALGITGPVLRATGLAHDLRRAQPYCGYENYDFDVVTHQDCDS
MMAR_1480|M.marinum_M YLDLAGCMALGITGPILRSTGLPHDLRKSEPYCGYENYEFDVITADGCDA
MUL_2462|M.ulcerans_Agy99 YLDLAGCMALGITGPILRSTGLPHDLRKSEPYCGYENYEFDVITADGCDA
Mb3172|M.bovis_AF2122/97 YLDLTGCMALGITGPILRSTGLPHDLRKSEPYCGYQHYEFDVITDDSCDA
Rv3148|M.tuberculosis_H37Rv YLDLTGCMALGITGPILRSTGLPHDLRKSEPYCGYQHYEFDVITDDSCDA
MAV_4036|M.avium_104 YLDLTGCMALGITGPILRATGLPHDLRKSEPYCGYENYEFDVITADTCDA
****:******:***:**::**.****:::*****: *:***:* : **:
MSMEG_2060|M.smegmatis_MC2_155 YGRYIIRVKEMHESVKIVEQCLARLKP---GPVMISDKKLAWPADLKLGP
TH_0634|M.thermoresistible__bu YGRFMIRVKEMHESLKIVEQCLDRLRP---GPVMIEDRKLAWPADLKLGP
Mflv_4484|M.gilvum_PYR-GCK YGRYLIRVKEMRESLRIVEQCVERLERSTGEPVMITDRKLAWPADLKVGP
Mvan_1882|M.vanbaalenii_PYR-1 YGRYLIRVKEMHQSIRIVEQCVQRLERSVGAPVMITDKKLAWPADLKVGP
MAB_2137|M.abscessus_ATCC_1997 YGRYLIRVKEMHESIKIAQQCVDRLRP---GPVMVDDKKIAWPADLASGP
MMAR_1480|M.marinum_M YGRYMIRVKEMWESIKIVEQCLDRLRP---GPTMIEDRKLAWPADLKVGP
MUL_2462|M.ulcerans_Agy99 YGRYMIRVKEMWESIKIVEQCLDRLRP---GPTMIEDRKLAWPADLKVGP
Mb3172|M.bovis_AF2122/97 YGRYMIRVKEMWESMKIVEQCLDKLRP---GPTMISDRKLAWPADLQVGP
Rv3148|M.tuberculosis_H37Rv YGRYMIRVKEMWESMKIVEQCLDKLRP---GPTMISDRKLAWPADLQVGP
MAV_4036|M.avium_104 YGRYMIRVKEMWESIKIVEQCLDKLQP---GPIMVENGKIAWPADLKVGP
***::****** :*::*.:**: :*. * *: : *:****** **
MSMEG_2060|M.smegmatis_MC2_155 DGLGNSPEHIAKIMGRSMEGLIHHFKLVTEGIRVPPGQVYVAVESPRGEL
TH_0634|M.thermoresistible__bu DGLGNSPEHVARIMGRSMEGLIHHFKLVTEGIRVPPGQVYVAVESPRGEL
Mflv_4484|M.gilvum_PYR-GCK DGLGNSPEHIAKIMGHSMEGLIHHFKLVTEGIRVPPGQVYVAVESPRGEL
Mvan_1882|M.vanbaalenii_PYR-1 DGLGNSPEHIAKIMGHSMEGLIHHFKLVTEGIRVPAGQVYVAVESPRGEL
MAB_2137|M.abscessus_ATCC_1997 DGLGNSPKHIAKIMGTSMEGLIHHFKLVTEGVRVPAGQVYVAIESPRGEL
MMAR_1480|M.marinum_M DGMGNSPEHIAKIMGSSMEALIHHFKLVTEGIRVPAGQVYVAVESPRGEL
MUL_2462|M.ulcerans_Agy99 DGMGNSPEHIAKIMGSSMEALIHHFKLVTEGIRVPAGQVYVAVESPRGEL
Mb3172|M.bovis_AF2122/97 DGLGNSPKHIAKIMGSSMEALIHHFKLVTEGIRVPAGQVYVAVESPRGEL
Rv3148|M.tuberculosis_H37Rv DGLGNSPKHIAKIMGSSMEALIHHFKLVTEGIRVPAGQVYVAVESPRGEL
MAV_4036|M.avium_104 DGLGNSPEHIAKIMGGSMEALIHHFKLVTEGIRVPPGQVYTAVESPRGEL
**:****:*:*:*** ***.***********:***.****.*:*******
MSMEG_2060|M.smegmatis_MC2_155 GVHMVSDGGTRPYRVHYRDPSFTNLQAVAATCEGGMVADAIAAVASIDPV
TH_0634|M.thermoresistible__bu GVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMAEGGMVADLIAALASIDPV
Mflv_4484|M.gilvum_PYR-GCK GVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMCEGGMVADAITAVASIDPV
Mvan_1882|M.vanbaalenii_PYR-1 GVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMCEGGMVADAITAVASIDPV
MAB_2137|M.abscessus_ATCC_1997 GVHMVSDGGTRPYRVHFRDPSFTNLQSVAAMCEGAMVADLIAAVASIDPV
MMAR_1480|M.marinum_M GVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMSEGGMVADLITAVASIDPV
MUL_2462|M.ulcerans_Agy99 GVHMVSDGGTRPYRVHYRDPSFTNLQAVAAMSEGGMVADLITAVASIDPV
Mb3172|M.bovis_AF2122/97 GVHMVSDGGTRPYRVHYRDPSFTNLQSVAAMCEGGMVADLIAAVASIDPV
Rv3148|M.tuberculosis_H37Rv GVHMVSDGGTRPYRVHYRDPSFTNLQSVAAMCEGGMVADLIAAVASIDPV
MAV_4036|M.avium_104 GVHMVSDGGTRPYRVHYRDPSFTNLQSVAAMCEGGMVADLITAVASIDPV
****************:*********:*** .**.**** *:*:******
MSMEG_2060|M.smegmatis_MC2_155 MGGVDR
TH_0634|M.thermoresistible__bu MGGVDR
Mflv_4484|M.gilvum_PYR-GCK MGGVDR
Mvan_1882|M.vanbaalenii_PYR-1 MGGVDR
MAB_2137|M.abscessus_ATCC_1997 MGGVDR
MMAR_1480|M.marinum_M MGGVDR
MUL_2462|M.ulcerans_Agy99 MGGVDR
Mb3172|M.bovis_AF2122/97 MGGVDR
Rv3148|M.tuberculosis_H37Rv MGGVDR
MAV_4036|M.avium_104 MGGVDR
******