For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RAVLIVNPTATSITPADRELVVNALKGRLDLDIEHTKHRGHATELGEAAAAAGVDLVVVHGGDGTVSGVI NGVLGRPGSTPTGPLPAIAVIPGGSANVLARSLGISHGPVAATNQLLQLLDDYRRRPQWRRIGLIDCGER WAAFNTGMGVDAEVVAAVEAERDGGRKVTPWRYVRATVPAMLSNAGREPALKLEIPGREPVTEVSFVFVS NASPWTFVGNRPVWTNPGCTFETGLGVFAMRNMKTIPTLRLLRQMLANRPKFDSKRLVRDDDVPSLRVTS TAAPVACQFDGDYLGVRQTMTFRAVPEAVAVVAPPPKNPC*
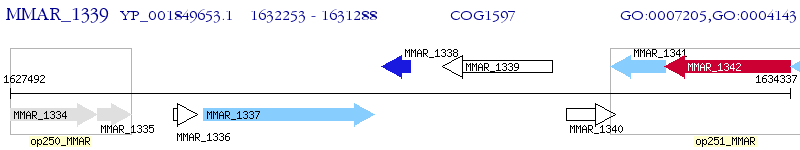
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1339 | - | - | 100% (321) | hypothetical protein MMAR_1339 |
| M. marinum M | MMAR_0569 | - | 2e-05 | 23.90% (318) | hypothetical protein MMAR_0569 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3244 | - | 1e-135 | 71.07% (318) | hypothetical protein Mb3244 |
| M. gilvum PYR-GCK | Mflv_4684 | - | 1e-106 | 59.12% (318) | diacylglycerol kinase, catalytic region |
| M. tuberculosis H37Rv | Rv3218 | - | 1e-135 | 71.07% (318) | hypothetical protein Rv3218 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3538 | - | 5e-97 | 54.37% (320) | hypothetical protein MAB_3538 |
| M. avium 104 | MAV_4166 | - | 1e-140 | 75.00% (316) | diacylglycerol kinase catalytic subunit |
| M. smegmatis MC2 155 | MSMEG_1920 | - | 1e-110 | 59.81% (321) | diacylglycerol kinase, catalytic region |
| M. thermoresistible (build 8) | TH_0670 | - | 1e-103 | 57.86% (318) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2540 | - | 0.0 | 98.44% (321) | hypothetical protein MUL_2540 |
| M. vanbaalenii PYR-1 | Mvan_1783 | - | 1e-108 | 60.06% (318) | diacylglycerol kinase, catalytic region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4684|M.gilvum_PYR-GCK MRAVLIVNPNATSTTPAGRDLLAHALESRVELSVVHTDHRGHAIEIAQDA
Mvan_1783|M.vanbaalenii_PYR-1 MRAVLIVNPNATSTTPAGRDLLAHALESRVRLTVTHTDHRGHAIEIAQQA
MSMEG_1920|M.smegmatis_MC2_155 MRAVLIVNPNATSTTAAGRDLLAHALESRVSLTVAHTDHRGHAIEIAREA
TH_0670|M.thermoresistible__bu VRAVLIVNPNATSTTPAGRDLLAYALESQTKLTVVHTAHRGHAIEIGRQA
MAB_3538|M.abscessus_ATCC_1997 MRAVLIVNPNATSTTAAGRDLLAHALESRVHLQVLHTDHRGHASELARRA
MMAR_1339|M.marinum_M MRAVLIVNPTATSITPADRELVVNALKGRLDLDIEHTKHRGHATELGEAA
MUL_2540|M.ulcerans_Agy99 MRAVLIVNPTATSITPADRVLVVNALKGRLDLDIEHTKHRGHATELGEAA
Mb3244|M.bovis_AF2122/97 MRAVLIVNPTATATTPAGRDLLAHALESRLQLTVEHTNHRGHGTELGQAA
Rv3218|M.tuberculosis_H37Rv MRAVLIVNPTATATTPAGRDLLAHALESRLQLTVEHTNHRGHGTELGQAA
MAV_4166|M.avium_104 MRAVLIVNPTATSTTAAGRDLLAHALKSRLQLAVEHTNHRGHGTELAQKA
:********.**: *.*.* *:. **:.: * : ** ****. *:.. *
Mflv_4684|M.gilvum_PYR-GCK ARGGVDVVIVHGGDGTVNEVVNGIVGQCGP---GNPAPAVGVVPGGSANV
Mvan_1783|M.vanbaalenii_PYR-1 ARSGVDVLIVHGGDGTVNEVVNGILGECGP---GGAGPAVGVVPGGSANV
MSMEG_1920|M.smegmatis_MC2_155 TRDGVDVLIVHGGDGTVNEVVNGVLSECGSRPDADSAPAVSVVPGGSANV
TH_0670|M.thermoresistible__bu CRDGVDVLIVHGGDGTVNEVVNGVLQFCGP---GGAGPAIGVVPGGSANV
MAB_3538|M.abscessus_ATCC_1997 ADEGAAVIVVHGGDGTVSEVVNGLLGQPGHP-APGPLPAVAVVPGGSANV
MMAR_1339|M.marinum_M AAAGVDLVVVHGGDGTVSGVINGVLGRPGST-PTGPLPAIAVIPGGSANV
MUL_2540|M.ulcerans_Agy99 AAAGVDLVVVHGGDGTVSGVINGVLGRPGST-PTGPLPAIAVIPGGSANV
Mb3244|M.bovis_AF2122/97 VADGVDLVVVHGGDGTVSAVVNGMLGRPGTT-PVRPVPAVAVVPGGSANV
Rv3218|M.tuberculosis_H37Rv VADGVDLVVVHGGDGTVSAVVNGMLGRPGTT-PVRPVPAVAVVPGGSANV
MAV_4166|M.avium_104 LADGVDLVVVHGGDGTVSGVVNGLLGRPGSL-PTGPVPAVAVVPGGSANV
*. :::********. *:**:: * . **:.*:*******
Mflv_4684|M.gilvum_PYR-GCK FARALGISPDPIEATNQLVDLLGAFRRGGS-WRRIGLMDCGERWGVFTAG
Mvan_1783|M.vanbaalenii_PYR-1 FARALGISADPIEATNQLVDLLGAYRQGRT-WRRIGLMDCGERWGIFTAG
MSMEG_1920|M.smegmatis_MC2_155 FARALGISPDPIEATNQLVDLLSDYRNGND-WRRIGLMDCGERWAVFTAG
TH_0670|M.thermoresistible__bu FARALGISPDPIEATKQLVELLGAHRRGQP-WRRIGLMKCDDRWAVFTAG
MAB_3538|M.abscessus_ATCC_1997 FARTLGISADPAQATNQVVDLLDEHRREQRRWRRISLGHCGERWFIFNAG
MMAR_1339|M.marinum_M LARSLGISHGPVAATNQLLQLLDDYRRRPQ-WRRIGLIDCGERWAAFNTG
MUL_2540|M.ulcerans_Agy99 LARSLGISHGPVAATNQLLQLLDDYRRRPQ-WRRIGLIDCGERWAAFNTG
Mb3244|M.bovis_AF2122/97 LARALGISADPIAATNQLIQLLDDYGRHQQ-WRRIGLIDCGERWAVFNAG
Rv3218|M.tuberculosis_H37Rv LARALGISADPIAATNQLIQLLDDYGRHQQ-WRRIGLIDCGERWAVFNAG
MAV_4166|M.avium_104 LARSLGISRDPVAATNQLIQLLDDYQRHHT-WRRIGLIDCGERWAVFNTG
:**:**** .* **:*:::**. . . ****.* .*.:** *.:*
Mflv_4684|M.gilvum_PYR-GCK MGVDGDVVAAVEAQRAKGRKVTASRYIRVAVREMLAATRKEPQLTLHLPG
Mvan_1783|M.vanbaalenii_PYR-1 MGVDGEVVAAVEAQRAKGRKVTASRYIRVAIREVLASARKEPALTLHLPD
MSMEG_1920|M.smegmatis_MC2_155 MGVDGDVVAAVEAQRAKGRKVTAGRYIRVAVREVLASVRREPSLTLHLPG
TH_0670|M.thermoresistible__bu MGVDGDVVAAVERQRAKGRKVTAGRYVRVAVREMLSNVFADPALTLHLPY
MAB_3538|M.abscessus_ATCC_1997 MGLDGEVVAAMEAHRNKGKQVTAGRYVARSVLAFFRAARRQPTLTVELPG
MMAR_1339|M.marinum_M MGVDAEVVAAVEAERDGGRKVTPWRYVRATVPAMLSNAGREPALKLEIPG
MUL_2540|M.ulcerans_Agy99 MGVDAEVVAAVEAERDGGRKVTPWRYVRATVPAMLSNAGRGPALKLEIPG
Mb3244|M.bovis_AF2122/97 MGVDAEVVAAVEAERDKGGKVTAWRYIRAAVRAVLACTRREPALTLQLPN
Rv3218|M.tuberculosis_H37Rv MGVDAEVVAAVEAERDKGGKVTAWRYIRAAVRAVLACTRREPALTLQLPN
MAV_4166|M.avium_104 MGVDAEVVAAVEAERDKGGKVTAWRYIRAAVPAVWGYTRREPMLTLELPG
**:*.:****:* .* * :**. **: :: . . * *.:.:*
Mflv_4684|M.gilvum_PYR-GCK REPVGGVHFAFVSNSSPWTYANSRPVWTNPDTTFETGLGVFATTSMNIWA
Mvan_1783|M.vanbaalenii_PYR-1 REPVSGVHFAFVSNASPWTYANARPVWTNPDTTFETGLGVFATTSMNVWA
MSMEG_1920|M.smegmatis_MC2_155 REPVSGVHFAFVSNSSPWTYANTRPVWTNPDTTFETGLGVFATTSMNVWS
TH_0670|M.thermoresistible__bu QEPIEGVHFAFVSNASPWTYANSRPVWTNPTTTFETGLGVFAITSMSVWA
MAB_3538|M.abscessus_ATCC_1997 HEPQAGIHFAFISNSNPWTYANERAIWTNPGTGFESGLGIFASKSMNVVA
MMAR_1339|M.marinum_M REPVTEVSFVFVSNASPWTFVGNRPVWTNPGCTFETGLGVFAMRNMKTIP
MUL_2540|M.ulcerans_Agy99 RESVTEVSFVFVSNASPWTFVGNRPVWTNPGCTFETGLGVFAMRNMKTIP
Mb3244|M.bovis_AF2122/97 RDPITGVHFVFVSNSSPWTYANNRPVWTNPDCRFESGLGVFATTSMKVVP
Rv3218|M.tuberculosis_H37Rv RDPITGVHFVFVSNSSPWTYANNRPVWTNPDCRFESGLGVFATTSMKVVP
MAV_4166|M.avium_104 REPITGVSFAFVSNSSPWTFANERPVWTNPGCSFESGLGVFAPTTLKPIP
::. : *.*:**:.***:.. *.:**** **:***:** .:. .
Mflv_4684|M.gilvum_PYR-GCK NLGLARQMN--SRKPRLDAKHLIRDDDLAWVEVTSD-TPVACQIDGDFLG
Mvan_1783|M.vanbaalenii_PYR-1 NLRLVRQMI--SRTPRLEAKHLIREDDLKWLKVTSD-TPVACQIDGDYIG
MSMEG_1920|M.smegmatis_MC2_155 NLRLVRQML--SRKPRIEGKHLIRDDDVPTVTVTSD-TPVACQIDGDYVG
TH_0670|M.thermoresistible__bu NLRLVRQMV--SKNPRIEASHLIRDDDLPWLRITGS-KPVSCQIDGDFVG
MAB_3538|M.abscessus_ATCC_1997 NLGLVRHMVGTPRADRPAAKHLLREDDIPWVRVRAS-APIATQVDGDFLG
MMAR_1339|M.marinum_M TLRLLRQML--ANRPKFDSKRLVRDDDVPSLRVTSTAAPVACQFDGDYLG
MUL_2540|M.ulcerans_Agy99 TLRLLRQML--AKRPKFDSKRLVRDDDVPSLRVTSTAAPVACQFDGDYLG
Mb3244|M.bovis_AF2122/97 TLRVVRQMF--AKQPKFEFNHVINNDDVACLRVTSMGPPIASQFDGDYLG
Rv3218|M.tuberculosis_H37Rv TLRVVRQMF--AKQPKFEFNHVINNDDVACLRVTSMGPPIASQFDGDYLG
MAV_4166|M.avium_104 TLRIVRQMF--AKRPAFDFKQLITEDDLPCLRVTSTGGPVACQFDGDYLG
.* : *:* .. .::: :**: : : . *:: *.***::*
Mflv_4684|M.gilvum_PYR-GCK PRDRMRFTAVPGALQVVAPPARMGR--
Mvan_1783|M.vanbaalenii_PYR-1 KRNMMQFTAVPNALAVVAPAPKMGR--
MSMEG_1920|M.smegmatis_MC2_155 LRESMSFTAVPAALKVVAPPCSRP---
TH_0670|M.thermoresistible__bu KREEMTFRSVPDAIDVVSPPNK-----
MAB_3538|M.abscessus_ATCC_1997 LRSDLTFTAVPDVLDVVAPAKPAGKQS
MMAR_1339|M.marinum_M VRQTMTFRAVPEAVAVVAPPPKNPC--
MUL_2540|M.ulcerans_Agy99 VRQTMTFRAVPEAVAVVAPPSKNPC--
Mb3244|M.bovis_AF2122/97 VRETMTFRAVPDALAVVAPPARKRI--
Rv3218|M.tuberculosis_H37Rv VRETMTFRAVPDALAVVAPPARKRI--
MAV_4166|M.avium_104 VRETMTFKSVPDALAVVAPPRQKRL--
*. : * :** .: **:*.