For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGDQVARIPSGGIRQLGPVNWALAKLAARSVRAPEMHLFTALGYRQYLFWTWAIYSGRLLHGRLPRIDTE LVILRVAHLRSCEYELQHHRRMARAAGLDAQAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIKNR SVTPATWEQLEIHLNRRRLIEFCLLATQYDALAATITALGVPLDNPR*
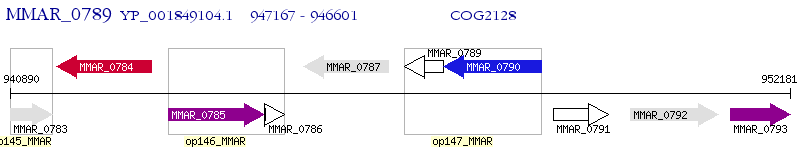
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0789 | - | - | 100% (188) | hypothetical protein MMAR_0789 |
| M. marinum M | MMAR_4743 | - | 9e-12 | 32.54% (126) | hypothetical protein MMAR_4743 |
| M. marinum M | MMAR_2345 | - | 3e-11 | 30.29% (175) | hypothetical protein MMAR_2345 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0473c | - | 1e-83 | 77.89% (190) | hypothetical protein Mb0473c |
| M. gilvum PYR-GCK | Mflv_0110 | - | 1e-49 | 53.30% (182) | carboxymuconolactone decarboxylase |
| M. tuberculosis H37Rv | Rv0464c | - | 1e-83 | 77.89% (190) | hypothetical protein Rv0464c |
| M. leprae Br4923 | MLBr_02465 | - | 4e-72 | 69.06% (181) | hypothetical protein MLBr_02465 |
| M. abscessus ATCC 19977 | MAB_4119 | - | 3e-49 | 52.78% (180) | hypothetical protein MAB_4119 |
| M. avium 104 | MAV_4685 | pcaC | 1e-67 | 67.78% (180) | hypothetical protein MAV_4685 |
| M. smegmatis MC2 155 | MSMEG_0905 | pcaC | 4e-48 | 53.55% (183) | hypothetical protein MSMEG_0905 |
| M. thermoresistible (build 8) | TH_0976 | - | 2e-40 | 53.95% (152) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4533 | - | 1e-107 | 99.47% (188) | hypothetical protein MUL_4533 |
| M. vanbaalenii PYR-1 | Mvan_0798 | - | 4e-50 | 54.40% (182) | carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0789|M.marinum_M -----MTG----DQVARIPSG-GIRQLGPVNWALAKLAARSVRAPEMHLF
MUL_4533|M.ulcerans_Agy99 -----MTG----DQVARIPSG-GIRQLGPVNWALAKLAARSVRAPEMHLF
Mb0473c|M.bovis_AF2122/97 -----MTGQN--GQVARISPG-KFRQLGPVNWLVAKLAARAVGAPQMHLF
Rv0464c|M.tuberculosis_H37Rv -----MTGQN--GQVARISPG-KFRQLGPVNWLVAKLAARAVGAPQMHLF
MLBr_02465|M.leprae_Br4923 -----MSGGTGTGPVGRIPPG-SLRQLGPINWVIAKLAASLLRTSEMHLF
MAV_4685|M.avium_104 -----MTA-------PRIPAG-RFRQLGPINWVIAKLGARTVGAPEMHLF
MAB_4119|M.abscessus_ATCC_1997 -----MSS-------GRIPSG-GLRELGPINWVIAKGMARAVNAPEMHLA
Mflv_0110|M.gilvum_PYR-GCK -----MS-------TARIPPG-GFKELGPLNWVIAKAGARGIRRPRFSLM
Mvan_0798|M.vanbaalenii_PYR-1 -----MSN------PARIAPG-GFKELGPLNWAIAKIGARGIRRPRFSLM
TH_0976|M.thermoresistible__bu ---MTATV------PPRIPPG-GFRELGPINWVIAKLGARGIRAPRFSLF
MSMEG_0905|M.smegmatis_MC2_155 MTRDHQTE------PARIPATRSLRALGPIGWIAARAGARAIRAPRFHLF
: **.. :: ***:.* *: * : ..: *
MMAR_0789|M.marinum_M TALGYRQYLFWTWAIYSGRLLHG-RLPRIDTELVILRVAHLRSCEYELQH
MUL_4533|M.ulcerans_Agy99 TALGYRQYLFWTWAIYSGRLLHG-RLPRIDTELVILRVAHLRSCEYELQH
Mb0473c|M.bovis_AF2122/97 TTLGYRQYLFWTFAIYTGRLLHG-RLPGVDTELVILRVAHLRSCEYELQH
Rv0464c|M.tuberculosis_H37Rv TTLGYRQYLFWTFAIYTGRLLHG-RLPGVDTELVILRVAHLRSCEYELQH
MLBr_02465|M.leprae_Br4923 TILGQRQLLFWAWLIYGGRLLRG-KLPRVDTELVILRVAHLRTCEYELQH
MAV_4685|M.avium_104 TTLGQRRLLFWTWLAYGGRLLRG-KLPTADTELVILRVAHLRGCEYELQH
MAB_4119|M.abscessus_ATCC_1997 TTLGQTGARFWPWLAYSGAILRGTKLSTRDTEVVILRVAHVRECEYELQH
Mflv_0110|M.gilvum_PYR-GCK NVLGQHRLLFLAWLPLSGFLLYAGKLSRRDAEVVILRVGHLRGSEYELQQ
Mvan_0798|M.vanbaalenii_PYR-1 NVLGQHRLLFLTWLPLSAHLLYVGKLSRHDAEVVILRVAHLRDSTYELQQ
TH_0976|M.thermoresistible__bu NVLGQHPLLFWTWLPYSGYLLYAGKLSRQDAEVVILRVGQLRDCEYELQQ
MSMEG_0905|M.smegmatis_MC2_155 DVIGQHKLAFLAFLPYSGVLLNWGKLPKRDTELVILRVAHLRGSTYELQQ
:* * .: . :* :*. *:*:*****.::* . ****:
MMAR_0789|M.marinum_M HRRMARAAGLDAQAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIK
MUL_4533|M.ulcerans_Agy99 HRRMARAAGLDAHAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIK
Mb0473c|M.bovis_AF2122/97 HRRMARRRGLDANTQATIFAWPDVPDGDGPRKVLSARQQALLQATDELIK
Rv0464c|M.tuberculosis_H37Rv HRRMARRRGLDANTQATIFAWPDVPDGDGPRKVLSARQQALLQATDELIK
MLBr_02465|M.leprae_Br4923 HRRMARKRGLDTKIQAMIFAWPDVPTGAG----LSVRQQALLAATDEFVK
MAV_4685|M.avium_104 HRRMARTAGLDPDLQAAIFAWPQRLEPVQAR--LTVRQQALLAATDEFVN
MAB_4119|M.abscessus_ATCC_1997 HTRIAKSAGIDPAYQERIFTG---AGAEG----LSDKERALITGVDEILT
Mflv_0110|M.gilvum_PYR-GCK HRRLARSRGLDKETQARIFAG---PDAEG----LTDRERVLITATDEFVV
Mvan_0798|M.vanbaalenii_PYR-1 HRRLARSRGVDADTQARIFAG---PDSEG----LTDRERVLITATDEFVV
TH_0976|M.thermoresistible__bu HRRLARSRGVGPELQAKIFEG---PDAEG----LTERQRALIAATDEFVL
MSMEG_0905|M.smegmatis_MC2_155 HRRLARSRGLDTETQAKIFEG---PDAEG----LTDRQRTLITATDEFVV
* *:*: *:. * ** *: :::.*: ..**::
MMAR_0789|M.marinum_M NRSVTPATWEQLEIHLNRRRLIEFCLLATQYDALAATITALGVPLDNPR-
MUL_4533|M.ulcerans_Agy99 NRSVTPATWEQLEIHLNRRRLIEFCLLATQYDALAATITALGVPLDNPR-
Mb0473c|M.bovis_AF2122/97 DRTITAGTWERLATHLDPRLLIEFCLLATQYDAIAATITALAIPPDNPQ-
Rv0464c|M.tuberculosis_H37Rv DRTITAGTWERLATHLDPRLLIEFCLLATQYDAIAATITALAIPPDNPQ-
MLBr_02465|M.leprae_Br4923 DRKITSSSWQQLETHLDRRRLIEFCMLISQYDGLAATISSLDIPLDNSC-
MAV_4685|M.avium_104 DRTVSEATWRQLAEHLDRRQLIEFCLLASQYDGLAATMSALAIPLDHPEG
MAB_4119|M.abscessus_ATCC_1997 TRTLSDEAWEGLSKFLDRRQLIGFCLLVTQYDGLAATMSSLRIPLDH---
Mflv_0110|M.gilvum_PYR-GCK TRGISPETWQMLAAHFTKPQLIEFCLLAAQYDGLAATISTLQVPLDFPD-
Mvan_0798|M.vanbaalenii_PYR-1 TRGVSPETWQALSAHLTKPQLIEFCLLAAQYDGLAATITTLQVPLDFPD-
TH_0976|M.thermoresistible__bu NRSVSSETWVRLAPPPPPPX---XXVGATPWPGGSVRRHDAR--------
MSMEG_0905|M.smegmatis_MC2_155 TRSMSPETWAALSTHLNRAQLIEFCLLAGHYDGLAATMATLRIPLDFPD-
* :: :* * : : . :.
MMAR_0789|M.marinum_M -
MUL_4533|M.ulcerans_Agy99 -
Mb0473c|M.bovis_AF2122/97 -
Rv0464c|M.tuberculosis_H37Rv -
MLBr_02465|M.leprae_Br4923 -
MAV_4685|M.avium_104 S
MAB_4119|M.abscessus_ATCC_1997 -
Mflv_0110|M.gilvum_PYR-GCK -
Mvan_0798|M.vanbaalenii_PYR-1 -
TH_0976|M.thermoresistible__bu -
MSMEG_0905|M.smegmatis_MC2_155 -