For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HSRLATARLYLCTDARRERGDLADFADAALAGGVDIIQLRDKGSAGEQRFGPLEARDELAACEILADAAA RHAAMFAVNDRADIARAARADVLHLGQRDLPVDVARAITGPATLIGQSTHDRDQVSAAAIGAVDYFCVGP CWPTPTKPGRTAPGLDLVRFAADVAGAKPWFAIGGIDGVRLPEVLAAGARRIVVVRAITAADDPREAAAK LKSELLAAI*
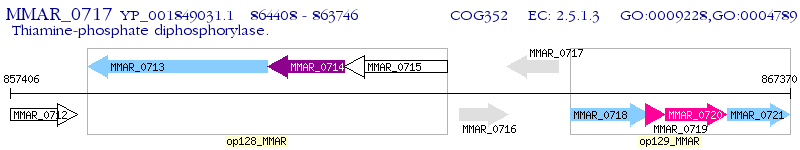
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0717 | thiE | - | 100% (220) | thiamine-phosphate pyrophosphorylase ThiE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0422c | thiE | 3e-95 | 80.73% (218) | thiamine-phosphate pyrophosphorylase |
| M. gilvum PYR-GCK | Mflv_0198 | thiE | 2e-82 | 72.22% (216) | thiamine-phosphate pyrophosphorylase |
| M. tuberculosis H37Rv | Rv0414c | thiE | 3e-95 | 80.73% (218) | thiamine-phosphate pyrophosphorylase |
| M. leprae Br4923 | MLBr_00300 | thiE | 1e-88 | 75.91% (220) | thiamine-phosphate pyrophosphorylase |
| M. abscessus ATCC 19977 | MAB_4219 | thiE | 3e-82 | 71.70% (212) | thiamine-phosphate pyrophosphorylase |
| M. avium 104 | MAV_4748 | thiE | 7e-98 | 80.18% (222) | thiamine-phosphate pyrophosphorylase |
| M. smegmatis MC2 155 | MSMEG_0789 | thiE | 8e-88 | 75.46% (216) | thiamine-phosphate pyrophosphorylase |
| M. thermoresistible (build 8) | TH_2311 | thiE | 8e-87 | 76.53% (213) | PROBABLE THIAMINE-PHOSPHATE PYROPHOSPHORYLASE THIE (TMP |
| M. ulcerans Agy99 | MUL_2806 | thiE | 1e-122 | 99.55% (220) | thiamine-phosphate pyrophosphorylase |
| M. vanbaalenii PYR-1 | Mvan_0707 | thiE | 4e-85 | 74.07% (216) | thiamine-phosphate pyrophosphorylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0717|M.marinum_M ----------------MHSR-----------LATARLYLCTDARRERGDL
MUL_2806|M.ulcerans_Agy99 ----------------MHSR-----------LATARLYLCTDARRERGDL
Mb0422c|M.bovis_AF2122/97 ----------------MHES----------RLASARLYLCTDARRERGDL
Rv0414c|M.tuberculosis_H37Rv ----------------MHES----------RLASARLYLCTDARRERGDL
MLBr_00300|M.leprae_Br4923 ----------------MQEPHRQPAICLSTRLAKARLYLCTDARRERGDL
MAV_4748|M.avium_104 ----------------MHQR--------LATLAAARLYLCTDARRERGDL
MSMEG_0789|M.smegmatis_MC2_155 -----------MDQAVELPV---------QRLQRASLYLCTDARRERGDL
TH_2311|M.thermoresistible__bu -------------------------------LSAASLYLCTDARRERGDL
Mflv_0198|M.gilvum_PYR-GCK MWSRSSLRRLTATVSTVPES---------PSLQGARLYLCTDARRERGDL
Mvan_0707|M.vanbaalenii_PYR-1 ----------------MRES---------RKLDSAALYLCTDARRERGDL
MAB_4219|M.abscessus_ATCC_1997 ----------------MHSR--------SARLQSAHLYLCTDARRERGDF
* * *************:
MMAR_0717|M.marinum_M ADFADAALAGGVDIIQLRDKGSAGEQRFGPLEARDELAACEILADAAARH
MUL_2806|M.ulcerans_Agy99 ADFADAALAGGVDIIQLRDKGSAGEQRFGPLEARDELAACEILADAAARH
Mb0422c|M.bovis_AF2122/97 AQFAEAALAGGVDIIQLRDKGSPGELRFGPLQARDELAACEILADAAHRY
Rv0414c|M.tuberculosis_H37Rv AQFAEAALAGGVDIIQLRDKGSPGELRFGPLQARDELAACEILADAAHRY
MLBr_00300|M.leprae_Br4923 AQFVNAALAGGVDIVQLRDKGSVGEQQFGPLEARDALAACEIFTDATGRH
MAV_4748|M.avium_104 AEFADAALAGGVDVIQLRDKGSPGEQRFGPLEARDELAACEILADAARRH
MSMEG_0789|M.smegmatis_MC2_155 AEFADAALAGGVDLIQLRDKGSAGEKQFGPLEARQELEALEILADAARRH
TH_2311|M.thermoresistible__bu AEFADAALAGGVDLIQLRDKGSPGEQRFGPLEARDELEALSVLADAARRH
Mflv_0198|M.gilvum_PYR-GCK ADFADAALAGGVDIIQLRDKGSPGEKAFGPLEARAEIDALHVLAEAARRH
Mvan_0707|M.vanbaalenii_PYR-1 AEFADAALAGGVDIIQLRDKGSAGEQRFGPLEAREEIEVLATLADAARRH
MAB_4219|M.abscessus_ATCC_1997 AEFVDAALAGGVDIVQLRDKGSAGERQFGRLEPAEELEYLAILSEAAARH
*:*.:********::******* ** ** *:. : :::*: *:
MMAR_0717|M.marinum_M AAMFAVNDRADIARAARADVLHLGQRDLPVDVARAITGPATLIGQSTHDR
MUL_2806|M.ulcerans_Agy99 AAMFAVNDRADIARAARADVLHLGQRDLPVDVARAITGPATLIGQSTHDR
Mb0422c|M.bovis_AF2122/97 GALFAVNDRADIARAAGADVLHLGQRDLPVNVARQILAPDTLIGRSTHDP
Rv0414c|M.tuberculosis_H37Rv GALFAVNDRADIARAAGADVLHLGQRDLPVNVARQILAPDTLIGRSTHDP
MLBr_00300|M.leprae_Br4923 DALFAVNDRADIARAAGADVLHLGQGDLPPGVARQIVCRQMLIGLSTHDR
MAV_4748|M.avium_104 GALFAVNDRADIARAAGADVLHLGQGDLPLDVARAFVGPDVLLGLSSHDR
MSMEG_0789|M.smegmatis_MC2_155 GALLAVNDRADIALAAGADVLHLGQDDLPLDVARGIIGRRPVIGRSTHDA
TH_2311|M.thermoresistible__bu DALLAVNDRADVALAAGADVLHLGQDDLPLPVARRIVGDEVLIGRSTHDI
Mflv_0198|M.gilvum_PYR-GCK NALVAVNDRADIARASGAEVLHLGQDDLPLTVARQIVGDR-VIGRSTHDL
Mvan_0707|M.vanbaalenii_PYR-1 GALFAVNDRADIALAADADVLHLGQDDLPLTVARRIVGDR-IVGRSTHDL
MAB_4219|M.abscessus_ATCC_1997 GALFAVNDRADIARAAGADVLHLGQDDLPLAVAREIVGPDVLIGRSTHDA
*:.*******:* *: *:****** *** *** : ::* *:**
MMAR_0717|M.marinum_M DQVSAAAIG----AVDYFCVGPCWPTPTKPGRTAPGLDLVRFAADVAGA-
MUL_2806|M.ulcerans_Agy99 DQVSAAAIG----AVDYFCVGPCWPTPTKPGRTAPGLDLVRFAADVAGA-
Mb0422c|M.bovis_AF2122/97 DQVAAAAAG-DA---DYFCVGPCWPTPTKPGRAAPGLGLVRVAAELGGDD
Rv0414c|M.tuberculosis_H37Rv DQVAAAAAG-DA---DYFCVGPCWPTPTKPGRAAPGLGLVRVAAELGGDD
MLBr_00300|M.leprae_Br4923 HQVAAAVAALDAGLVDYFCVGPCWPTPTKPDRPAPGLELVRAAAELAGD-
MAV_4748|M.avium_104 DQMTAAAAG----PADYFCVGPCWPTPTKPGRAAPGLALVRAAAELRTG-
MSMEG_0789|M.smegmatis_MC2_155 AQMAVAIEER----VDYFCVGPCWPTPTKPGRPAPGLDLVRATAAHSPG-
TH_2311|M.thermoresistible__bu EQVRTAIADRD---VDYFCVGPCWPTPTKPGRPAPGLDLVRAAAAEGTG-
Mflv_0198|M.gilvum_PYR-GCK AQAEAAIAED----VDYFCVGPCWPTPTKPGRAAPGLDLVREVAALNTT-
Mvan_0707|M.vanbaalenii_PYR-1 DQVRAAVGED----VNYFCVGPCWPTPTKPGRPAPGLDLIRATAALGTD-
MAB_4219|M.abscessus_ATCC_1997 EQAAAAARDDE---IDYFCCGPCWPTPTKPGRTASGLGLVRTAAELDTS-
* .* :*** **********.*.*.** *:* .*
MMAR_0717|M.marinum_M KPWFAIGGIDGVRLPEVLAAGARRIVVVRAITAADDPREAAAKLKSELLA
MUL_2806|M.ulcerans_Agy99 KPWFAIGGIDGLRLPEVLAAGARRIVVVRAITAADDPREAAAKLKSELLA
Mb0422c|M.bovis_AF2122/97 KPWFAIGGINAQRLPAVLDAGARRIVVVRAITSADDPRAAAEQLRSALTA
Rv0414c|M.tuberculosis_H37Rv KPWFAIGGINAQRLPAVLDAGARRIVVVRAITSADDPRAAAEQLRSALTA
MLBr_00300|M.leprae_Br4923 KPWFAIGGIDAQRLPDVLHAGARRIVVVRAITAAADPRAAAEQLISTLTA
MAV_4748|M.avium_104 KPWFAIGGIDAQRLPEVLDAGARRVVVVRAITAADDPAAAARRLSSALAA
MSMEG_0789|M.smegmatis_MC2_155 KPWFAIGGIDQERLPEVLAAGARRVVVVRAITAADDPKAAAEDLKAAISA
TH_2311|M.thermoresistible__bu KPWFAIGGIDEQRLPEVVAAGARRIVVVRAITAADDPEAAARRLKAMLPG
Mflv_0198|M.gilvum_PYR-GCK KPWFAIGGIDAQRLPDVLDAGARRVVVVRAITAAEDPRAAAEQLAGMLDS
Mvan_0707|M.vanbaalenii_PYR-1 KPWFAIGGIDAERLPEVLDAGARRVVVVRAITAADDPGAAAQRLAGMLSA
MAB_4219|M.abscessus_ATCC_1997 KPWFAIGGIDEARVPQVVEAGASRIVVVRAITAAADPRAAAASLRGSTRR
*********: *:* *: *** *:*******:* ** ** * .
MMAR_0717|M.marinum_M AI--
MUL_2806|M.ulcerans_Agy99 AI--
Mb0422c|M.bovis_AF2122/97 AN--
Rv0414c|M.tuberculosis_H37Rv AN--
MLBr_00300|M.leprae_Br4923 TS--
MAV_4748|M.avium_104 AR--
MSMEG_0789|M.smegmatis_MC2_155 AG--
TH_2311|M.thermoresistible__bu GRTG
Mflv_0198|M.gilvum_PYR-GCK AV--
Mvan_0707|M.vanbaalenii_PYR-1 AG--
MAB_4219|M.abscessus_ATCC_1997 A---