For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLRVVPEGLAAASAAVEALTARLAAAHASASPVISAVVPAAVDPVSLQTAAGFSAQGIEHAAVAAEGVE ELGRAGLGVGESGVSYLAGDAVAAATYGIAGA
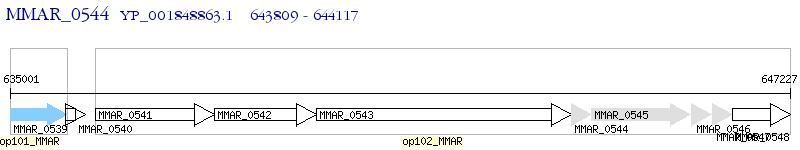
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0544 | - | - | 100% (102) | PE family protein, PE5 |
| M. marinum M | MMAR_3003 | - | 8e-36 | 70.71% (99) | PE family protein |
| M. marinum M | MMAR_5258 | - | 2e-35 | 69.70% (99) | PE family protein, PE5_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0293 | PE5 | 3e-45 | 88.12% (101) | PE family protein |
| M. gilvum PYR-GCK | Mflv_0326 | - | 9e-36 | 70.00% (100) | PE domain-containing protein |
| M. tuberculosis H37Rv | Rv0285 | PE5 | 3e-45 | 88.12% (101) | PE family protein |
| M. leprae Br4923 | MLBr_02534 | - | 5e-35 | 72.73% (99) | PE-family protein |
| M. abscessus ATCC 19977 | MAB_2231c | - | 2e-36 | 76.00% (100) | PE family protein |
| M. avium 104 | MAV_4868 | - | 1e-42 | 83.33% (102) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0618 | - | 1e-36 | 77.32% (97) | PE family protein |
| M. thermoresistible (build 8) | TH_3343 | - | 4e-08 | 42.25% (71) | PUTATIVE PE domain protein |
| M. ulcerans Agy99 | MUL_1207 | - | 2e-50 | 100.00% (102) | PE family protein |
| M. vanbaalenii PYR-1 | Mvan_0414 | - | 2e-34 | 70.00% (100) | PE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0544|M.marinum_M MTLRVVPEGLAAASAAVEALTARLAAAHASASPVISAVVPAAVDPVSLQT
MUL_1207|M.ulcerans_Agy99 MTLRVVPEGLAAASAAVEALTARLAAAHASASPVISAVVPAAVDPVSLQT
Mb0293|M.bovis_AF2122/97 MTLRVVPEGLAAASAAVEALTARLAAAHASAAPVITAVVPPAADPVSLQT
Rv0285|M.tuberculosis_H37Rv MTLRVVPEGLAAASAAVEALTARLAAAHASAAPVITAVVPPAADPVSLQT
MAV_4868|M.avium_104 MTLRVVPEGLAATSAAVEALTARLAAAHAAAAPAITAVVPPAADPVSLQT
Mflv_0326|M.gilvum_PYR-GCK MTLRVVPEGLTAASAAVEALTARMAAAHAAAAPLVTAVLPPAADAVSLQT
Mvan_0414|M.vanbaalenii_PYR-1 MTLRVVPEGLTAAGAAVEALTARMAAAHAAAAPVVTAVLPPAADVVSLQA
MSMEG_0618|M.smegmatis_MC2_155 MTLRVVPEGLTAASSAVEALTARLAAAHAAAAPMISTVLPPAADAVSLQT
MAB_2231c|M.abscessus_ATCC_199 MNLNVVPEGLTATSAAVEALTARLAAAHAAAAPVIGAVVPPAADPVSLET
MLBr_02534|M.leprae_Br4923 MTLRVVPEKLAATSEAMKALTARLEAAHAAAFPCLVAVVPPAADPVSLQT
TH_3343|M.thermoresistible__bu ----------VGIGSQVIANGTRGLAVGTAASAEVTALVPAGADEVSAQA
.. . : * :* *. ::* . : :::*...* ** ::
MMAR_0544|M.marinum_M AAGFSAQGIEHAAVAAEGVEELGRAGLGVGESGVSYLAGDAVAAATYGIA
MUL_1207|M.ulcerans_Agy99 AAGFSAQGIEHAAVAAEGVEELGRAGLGVGESGVSYLAGDAVAAATYGIA
Mb0293|M.bovis_AF2122/97 AAGFSAQGVEHAVVTAEGVEELGRAGVGVGESGASYLAGDAAAAATYGVV
Rv0285|M.tuberculosis_H37Rv AAGFSAQGVEHAVVTAEGVEELGRAGVGVGESGASYLAGDAAAAATYGVV
MAV_4868|M.avium_104 ALGFSAQGQEHSAVAAQGVEELGRAGVGVGEAGASYLAGDTAAAATFGIA
Mflv_0326|M.gilvum_PYR-GCK ATGLSANGVEHQSIAARGVEELGRSGVGVGDSAASYVSGDAMAATSYLIA
Mvan_0414|M.vanbaalenii_PYR-1 ATGFSARAAEHQSVAAQGVEELGRSGIAVGESAASYAGGDAMAASSYLIA
MSMEG_0618|M.smegmatis_MC2_155 AAGFSANGAQQSAVAAQGVEELGRSGVGVGESGVSYATGDAQAAASYLTA
MAB_2231c|M.abscessus_ATCC_199 AAGFSARGIEHSGVAAQAVEELGRAGLGVSESAASYTTGDMQAAAAYMIA
MLBr_02534|M.leprae_Br4923 AAGFSARGQEHALVAAQGVEELGRAGIGVGQSSTHYAISDALAASTYGIV
TH_3343|M.thermoresistible__bu AAAFAKEGMEALALNTFAQEELARAGAAVVQIAGIYDAVDAANAGTLA--
* .:: .. : : : . ***.*:* .* : . * * * :
MMAR_0544|M.marinum_M GA-
MUL_1207|M.ulcerans_Agy99 GA-
Mb0293|M.bovis_AF2122/97 GG-
Rv0285|M.tuberculosis_H37Rv GG-
MAV_4868|M.avium_104 GA-
Mflv_0326|M.gilvum_PYR-GCK RG-
Mvan_0414|M.vanbaalenii_PYR-1 RG-
MSMEG_0618|M.smegmatis_MC2_155 RGL
MAB_2231c|M.abscessus_ATCC_199 RG-
MLBr_02534|M.leprae_Br4923 ES-
TH_3343|M.thermoresistible__bu ---