For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTELDEVDSLRSDGDSWTVTESVGATALGVAAARAVETAGANPLIRDEFAPILVSSAGPAWARLADPDIG WLDDDPHGQRLHRLGCDYQAVRTHFFDEYFAAAAGAGIEQAVILAAGLDCRAYRLNWPPEAVVFEIDQPK VLEYKAQILESHGVTAAATRHGVAVDLREDWPAALLRAGFDRDRPTAWLAEGLLPYLPGDAQDRLFEMIT DLSAPRSRIAVESFTMNLTGNKQRWNRMRDRLGLDINVEALTYREPGRADAAEWLANHGWQVYSVSNREE MARLGRPVPEDLVDEAITTTLLRASLEISR
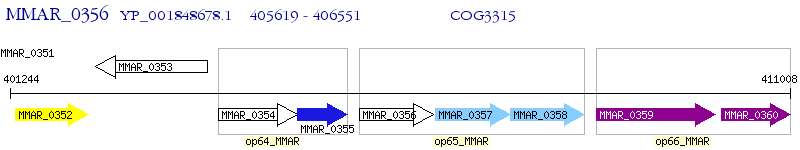
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0356 | - | - | 100% (310) | hypothetical protein MMAR_0356 |
| M. marinum M | MMAR_0357 | - | 3e-67 | 48.15% (297) | O-methyltransferase |
| M. marinum M | MMAR_5323 | - | 2e-66 | 47.65% (298) | O-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0150 | - | 1e-134 | 73.87% (310) | hypothetical protein Mb0150 |
| M. gilvum PYR-GCK | Mflv_0743 | - | 4e-75 | 51.03% (290) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv0145 | - | 1e-134 | 73.87% (310) | hypothetical protein Rv0145 |
| M. leprae Br4923 | MLBr_02640 | - | 1e-64 | 48.80% (291) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_4607c | - | 8e-84 | 54.22% (308) | hypothetical protein MAB_4607c |
| M. avium 104 | MAV_5150 | - | 1e-128 | 72.40% (308) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_0093 | - | 1e-99 | 58.93% (319) | methyltransferase |
| M. thermoresistible (build 8) | TH_2759 | - | 1e-104 | 60.83% (314) | PUTATIVE methyltransferase |
| M. ulcerans Agy99 | MUL_4763 | - | 1e-179 | 99.03% (310) | hypothetical protein MUL_4763 |
| M. vanbaalenii PYR-1 | Mvan_0104 | - | 4e-77 | 53.24% (293) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0356|M.marinum_M ---MTELDEVDSLRSD----GDSWTVTESVGATALGVAAARAVETAGANP
MUL_4763|M.ulcerans_Agy99 ---MTELDEVDSLRSD----GDSWTVTESVGATALGVAAARAVETAGANP
Mb0150|M.bovis_AF2122/97 ---MTELDDVSSLPSSRRTAGDTWAITESVGATALGVAAARAVETAATNP
Rv0145|M.tuberculosis_H37Rv ---MTELDDVSSLPSSRRTAGDTWAITESVGATALGVAAARAVETAATNP
MAV_5150|M.avium_104 ---MTDLD--SSLPPSVRTAGDSWTITELVGATALGVAAARAAETAGPDP
MSMEG_0093|M.smegmatis_MC2_155 ---MTDVRETPSVRSA----RDSWGITESVGATALGVAAARAAETAQADP
TH_2759|M.thermoresistible__bu MVMMFDVTEAASARSE----GDSWDITESVGATALGVAAERALETSQPDP
MAB_4607c|M.abscessus_ATCC_199 ------------MRVD----GDTWDITSSVGATALGVAAARATETLRADA
Mflv_0743|M.gilvum_PYR-GCK ---------MSSLRTD----DDTWDIASSVGATAVMVAAARAAETERHDA
Mvan_0104|M.vanbaalenii_PYR-1 ---------MSSLRTD----NDTWDIASSVGATAVMVAAARAAETERPEP
MLBr_02640|M.leprae_Br4923 ------------MRTH----DDTWDIKTSVGTTAVMVAAARAAETDRPDA
*:* : **:**: *** ** ** :.
MMAR_0356|M.marinum_M LIRDEFAPILVSSAG--PAWARLADP--DIGWLDDDPHGQRLHRLGCDYQ
MUL_4763|M.ulcerans_Agy99 LIRDEFAPILVSSAG--PAWARLADP--DIGWLDDDPHGQRLHRLGCDYQ
Mb0150|M.bovis_AF2122/97 LIRDEFAKVLVSSAG--TAWARLADA--DLAWLDGDQLGRRVHRVACDYQ
Rv0145|M.tuberculosis_H37Rv LIRDEFAKVLVSSAG--TAWARLADA--DLAWLDGDQLGRRVHRVACDYQ
MAV_5150|M.avium_104 LIRDEFAGLLVSSAG--PAWARLADP--EQSWLDDDPHGKRAHRVGIDYQ
MSMEG_0093|M.smegmatis_MC2_155 LIRDEFAFLLVSAAG--PTWAQMAVS--DPHWLGADADARRIHEISRNYQ
TH_2759|M.thermoresistible__bu LIRDEYAFLLVSAAG--PPWTQLASS--DLSWLGDDETARRMHLMSRDYQ
MAB_4607c|M.abscessus_ATCC_199 LIRDPFAQILVDATGKATGWERLVAG--DIDWP--DPEAGRIYDRMVDYQ
Mflv_0743|M.gilvum_PYR-GCK LINDPYAKVLVEGAGAGA-WRFIADEDLVAKASESDTEVGALFEHMKNYQ
Mvan_0104|M.vanbaalenii_PYR-1 LIDDPYAKILVAGAGNGA-WQYIADDGFVAKVTESDPEIGPLFEHMKNYQ
MLBr_02640|M.leprae_Br4923 LIRDPYAKLLVTNTGAGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQ
** * :* :** :* . * : * .**
MMAR_0356|M.marinum_M AVRTHFFDEYFAAAAGAGIEQAVILAAGLDCRAYRLNWPPEAVVFEIDQP
MUL_4763|M.ulcerans_Agy99 AVRTHFSDEYFAAAAGAGIEQAVILAAGLDCRAYRLNWPPEAVVFEIDQP
Mb0150|M.bovis_AF2122/97 AVRTHFFDEYFGAAVDAGVRQVVILAAGLDARAYRLNWPAGTVVYEIDQP
Rv0145|M.tuberculosis_H37Rv AVRTHFFDEYFGAAVDAGVRQVVILAAGLDARAYRLNWPAGTVVYEIDQP
MAV_5150|M.avium_104 AVRTHYFDEYFDGALRAGIRQVVILAAGLDSRAYRLNWPAGTTVYEIDQP
MSMEG_0093|M.smegmatis_MC2_155 AVRTHYFDEYFSDVAHAGIRQVVILAAGLDSRAFRLDWPAGTTVFEIDQP
TH_2759|M.thermoresistible__bu AVRTHYFDRYFAAAARAGIRQIVLLAAGLDSRAYRLDWPAGTVVYELDQP
MAB_4607c|M.abscessus_ATCC_199 ATRTHFFDEYFLAAAAAGIRQVVILASGLDSRAYRLDWPAGTTVYEIDQP
Mflv_0743|M.gilvum_PYR-GCK AVRTHFFDAFFSAAVDAGVRQIVILASGLDSRAFRLPWPAGTVVYEIDQP
Mvan_0104|M.vanbaalenii_PYR-1 AVRTHFFDAFFAAAVDAGIRQIVILASGLDSRAFRLPWPAGTTVFEIDQP
MLBr_02640|M.leprae_Br4923 AVRTNFFDTYFNNAVIDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQP
*.**:: * :* . *:.* *:**:***.**:** **. :.*:*:***
MMAR_0356|M.marinum_M KVLEYKAQILESHGVTAAATRHGVAVDLREDWPAALLRAGFDRDRPTAWL
MUL_4763|M.ulcerans_Agy99 KVLEYKAQILESHGVTAAATRHGVAVDLREDWPAALLRAGFDRDRPTAWL
Mb0150|M.bovis_AF2122/97 SVLEYKAGILQSHGAVPTARRHAVAVDLRDDWPAALIAAGFDGTQPTAWL
Rv0145|M.tuberculosis_H37Rv SVLEYKAGILQSHGAVPTARRHAVAVDLRDDWPAALIAAGFDGTQPTAWL
MAV_5150|M.avium_104 KVLEYKTETLQRHGATPAAVRRPVPVDLRDDWPAALTAAGFQAARPTAWL
MSMEG_0093|M.smegmatis_MC2_155 KVLEYKTATLDAHGAVAKARYVPVPADLRDDWPAALVEAGFDPAQPTAWL
TH_2759|M.thermoresistible__bu RVLDFKNRTLQAHGAVAKADRRTVAVDLRDDWPAALRHAGFDSAAPTAWL
MAB_4607c|M.abscessus_ATCC_199 QVLKFKDSALAAH--QPTAQRRGVAIDLREDWPAALRAAGFDSAQPTAWL
Mflv_0743|M.gilvum_PYR-GCK LVLDYKSSTLAAHGVAPTAERREVPIDLRQDWPAALVATGFDPARPTAWL
Mvan_0104|M.vanbaalenii_PYR-1 LVLAYKSSTLASHGVQPTADRREVPIDLRQDWPTALTHAGFDADQPTAWL
MLBr_02640|M.leprae_Br4923 KVLAYKSTTLAEHGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWL
** :* * * . * *. ***:***.** :**: ****
MMAR_0356|M.marinum_M AEGLLPYLPGDAQDRLFEMITDLSAPRSRIAVESFTMNLTG-NKQ-----
MUL_4763|M.ulcerans_Agy99 AEGLLPYLPGDAQDRLFEMITDLSAPRSRIAVESFTMNLTG-NKQ-----
Mb0150|M.bovis_AF2122/97 AEGLLPYLPGDAADRLFDMVTALSAPGSQVAVEAFTMNTKG-NTQ-----
Rv0145|M.tuberculosis_H37Rv AEGLLPYLPGDAADRLFDMVTALSAPGSQVAVEAFTMNTKG-NTQ-----
MAV_5150|M.avium_104 AEGLLPYLPSDAQDRLFEMVTALSAAGSQVAVEVFGMNSRS-NAQ-----
MSMEG_0093|M.smegmatis_MC2_155 AEGLLPYLPADAQDRLFELVGVNSAPGSRIAVEAFNMSPER-YTEERRAQ
TH_2759|M.thermoresistible__bu AEGLLCYLPADAQDRLFERITALSAPGSWLAVEAFHHDPQR-ATERRRAE
MAB_4607c|M.abscessus_ATCC_199 AEGLLPYLPADAQDNLFRDIGVLSAAGSRVAVEGYEGKLVLDESEEEAVR
Mflv_0743|M.gilvum_PYR-GCK AEGLLMYLPADAQDRLFAQITELSAPGSQVAAESMGVHAPDRRER-----
Mvan_0104|M.vanbaalenii_PYR-1 AEGLLMYLPADAQDRLFAQITELSAPGSRVAAESMGIHAQDRRER-----
MLBr_02640|M.leprae_Br4923 AEGLLMYLPATAQDGLFTEIGGLSAVGSRIAVETSPLHGDEWREQ-----
***** ***. * * ** : ** * :*.* .
MMAR_0356|M.marinum_M ---RWNRMRDRLGLDI--NVEALTYREPGRADAAEWLANHGWQVYSVSNR
MUL_4763|M.ulcerans_Agy99 ---RWNRMRDRLGLDI--NVEALTYREPGRTDAAEWLANHGWQVYSVSNR
Mb0150|M.bovis_AF2122/97 ---RWNRMRERLGLDI--DVQALTYHEPDRSDAAQWLATHGWQVHSVSNR
Rv0145|M.tuberculosis_H37Rv ---RWNRMRERLGLDI--DVQALTYHEPDRSDAAQWLATHGWQVHSVSNR
MAV_5150|M.avium_104 ---RWLRMRERLGLDV--NVAALTYHEPDRSDAAAWLTGHGWRVHSVDNR
MSMEG_0093|M.smegmatis_MC2_155 RRARGEQMRKQLDLDI--DVDALMYTADDRADAAQWLSEHGWQVEAVPSA
TH_2759|M.thermoresistible__bu WRDRTARMKAALDLNL--DFEALTYDDPDRADAGEWLAAHGWHPDSVPGT
MAB_4607c|M.abscessus_ATCC_199 REQVRAAFKTAIDVDV--TIENLIYEDENRADPAEWLDAHGWSVTKTDAH
Mflv_0743|M.gilvum_PYR-GCK MRERFASIASQIDIEP-MDITELTYEDPDRAEVAEWLAAHGWTAQAVPSQ
Mvan_0104|M.vanbaalenii_PYR-1 MRERFASITAQFDVEP-MDITELTYEDPDRADVAQWLTAHGWRAEAVPSQ
MLBr_02640|M.leprae_Br4923 MQLRFRRVSDALGFEQAVDVQELIYHDENRAVVADWLNRHGWRATAQSAP
. :..: . * * .*: . ** ***
MMAR_0356|M.marinum_M EEMARLGRP---VPEDLVDEAITTTLLRASLEISR---
MUL_4763|M.ulcerans_Agy99 EEMARLGRP---VPEDLVDEAITTTLLRASLEILR---
Mb0150|M.bovis_AF2122/97 EEMARLGRA---IPQDLVDETVRTTLLRGRLVTPAQPA
Rv0145|M.tuberculosis_H37Rv EEMARLGRA---IPQDLVDETVRTTLLRGRLVTPAQPA
MAV_5150|M.avium_104 DEMARLGRP---VPEDLSDEAVRSTLLRAHLGGSTG--
MSMEG_0093|M.smegmatis_MC2_155 DEMARLGRPA--AEADLAEFGMDGVLMRARLDGERL--
TH_2759|M.thermoresistible__bu EEMARMGRP---VPADLLDVAVPSTLLRAHLNGASR--
MAB_4607c|M.abscessus_ATCC_199 DEMARLGRP---VAEDVERGTFRGQLIQGELR------
Mflv_0743|M.gilvum_PYR-GCK DAMRRLDRY---VEVADSDGQSFSTFTTGTKL------
Mvan_0104|M.vanbaalenii_PYR-1 DEMRRLHRL---VEIADGDDQSFSTFTTAVKR------
MLBr_02640|M.leprae_Br4923 DEMRRVGRWGDGVPMAD-DKDAFAEFVTAHRL------
: * *: * : .