For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
INDLLIRWLATGLFVFSGADYALSALSALSALPALPALPALPALSTFSALPASSAFSGAPTVTQQRAWTS VVSNGLHLVMAIAMVVMLWPVGAALPTTGPAVFFLLAAGWFATLTALRAKTITALVGCSYHALMMLAMAW MYVAMSDQLLACPAAHQTMAPGTSMPGMHMSGTDMAAGATAPGWVSAVNWFWLLAFAIAAIIWAVMSVRA RPRAGTESWRSCLTHAGRAMMAAGMAIAFAAMVFGI*
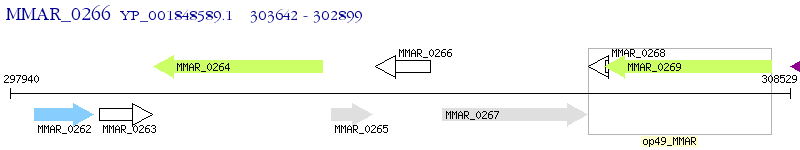
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0266 | - | - | 100% (247) | hypothetical protein MMAR_0266 |
| M. marinum M | MMAR_1432 | - | 3e-43 | 41.94% (248) | hypothetical protein MMAR_1432 |
| M. marinum M | MMAR_4877 | - | 2e-37 | 37.90% (248) | hypothetical protein MMAR_4877 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0995 | - | 3e-44 | 40.40% (250) | integral membrane protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0970 | - | 3e-44 | 40.40% (250) | integral membrane protein |
| M. leprae Br4923 | MLBr_01997 | - | 1e-41 | 39.11% (248) | putative integral membrane protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0088 | - | 1e-27 | 31.30% (246) | hypothetical protein MAV_0088 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4848 | - | 1e-136 | 97.98% (247) | hypothetical protein MUL_4848 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0266|M.marinum_M MINDLLIRWLATGLFVFSGADYALSALSALSALPALPALPALPALSTFSA
MUL_4848|M.ulcerans_Agy99 MINDLLIRWLATGLFVFSGVDYALSALSALSALPALPALPALPALSTFSA
Mb0995|M.bovis_AF2122/97 MIHDLMLRWVVTGLFVLTAAECGLAIIAKR--------------------
Rv0970|M.tuberculosis_H37Rv MIHDLMLRWVVTGLFVLTAAECGLAIIAKR--------------------
MLBr_01997|M.leprae_Br4923 MIDDLLLRWVVTGLFVLSAAECGFACLACR--------------------
MAV_0088|M.avium_104 MIGELTVRWAVTALFGASLAAYAYLAVSQR--------------------
** :* :** .*.** : . . ::
MMAR_0266|M.marinum_M LPASSAFSGAPTVTQQRAWTSVVSNGLHLVMAIAMVVMLWPVGAALPTTG
MUL_4848|M.ulcerans_Agy99 LPASSAFSGAPTVTQQRAWTSVVSNGLHLVMAIAMVVMLWPVGAALPTTG
Mb0995|M.bovis_AF2122/97 ----------------RPWTLIVNHGLHFAMAVAMAVMAWPWGARVPTTG
Rv0970|M.tuberculosis_H37Rv ----------------RPWTLIVNHGLHFAMAVAMAVMAWPWGARVPTTG
MLBr_01997|M.leprae_Br4923 ----------------RPWTLVPSNGLHFAMAIAMAVMAWPRGAQLPTTG
MAV_0088|M.avium_104 ----------------GRWSCAVNHLLHLAMSAAMILMVWRVGLGLPAVA
*: .: **:.*: ** :* * * :*:..
MMAR_0266|M.marinum_M PAVFFLLAAGWFATLTALRAKTITALVGCSYHALMMLAMAWMYVAMSDQL
MUL_4848|M.ulcerans_Agy99 PAVFFLLAAVWFATLTALRAKTTTALVGCSYHALMMLAMAWMYVAMSDQL
Mb0995|M.bovis_AF2122/97 PAVFFLLAAVWFGATAVVAVRGTATRGLYGYHGLMMLATAWMYAAMNPRL
Rv0970|M.tuberculosis_H37Rv PAVFFLLAAVWFGATAVVAVRGTATRGLYGYHGLMMLATAWMYAAMNPRL
MLBr_01997|M.leprae_Br4923 TVVFCGLVGAWFVILATVSSRRIAERAAYAYPALMMLAMVWMYVIMDS-H
MAV_0088|M.avium_104 PTLFFLLAGAWFVFLAVRASAESRERLKTCYYAAMMAAMAWMYALMSSGT
..:* *.. ** :. * . ** * .***. *.
MMAR_0266|M.marinum_M LACP-AAHQTMA-PGTSMPGMHMSGTDMAAGATAPGWVSAVNWFWLLAFA
MUL_4848|M.ulcerans_Agy99 LACP-AAHQTMA-PGTSMPGMHMSGTDMAAGATAPGWVSAVNWFWLLAFA
Mb0995|M.bovis_AF2122/97 LPVRSCTEYATE-PDGSMPAMDMTAMNMPPNSGSPIWFSAVNWIGTVGFA
Rv0970|M.tuberculosis_H37Rv LPVRSCTEYATE-PDGSMPAMDMTAMNMPPNSGSPIWFSAVNWIGTVGFA
MLBr_01997|M.leprae_Br4923 LHDHWATGHHTS-PHTSMLGVDMTTT-VVPASGIPGWISIINWLWFAFFC
MAV_0088|M.avium_104 TGAHAHGHSASDSVGANMPGMRLPEHPASPGAGGFSWVAAANWLGALGFA
.* .: :. . : *.: **: *.
MMAR_0266|M.marinum_M IAAIIWAVMSVRARPRAGTESWRSCLTHAGRAMMAAGMAIAFAAMVFGI
MUL_4848|M.ulcerans_Agy99 IAAIIRAAMSVRARPRAGTESWRSCLTHAGRAMMAAGMAIAFAAMVFGI
Mb0995|M.bovis_AF2122/97 VAAVFWACRFVMERRQEATQSRLP--GSIGQAMMAAGMAMLFFAMLFPV
Rv0970|M.tuberculosis_H37Rv VAAVFWACRFVMERRQEATQSRLP--GSIGQAMMAAGMAMLFFAMLFPV
MLBr_01997|M.leprae_Br4923 IATVFWTYRSFATSRRNAGFSRYCSLHPSGQAMMSTGMAIMFGVLLFHV
MAV_0088|M.avium_104 ALAVYWLYRFVGQRRLARVPARLARMEPLYQAFTAAGTALMFDTLLR--
:: . : :*: ::* *: * .::