For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARETARILRMLSAVALGSLVVWTGGCSAGEHAAGAPAPATIPVGRSAQSIQWDGAERTFNLYRPQGLTGA VPLVVMMHGGFGNGAQAERAYHWNDKADQQHFLVAYPDGLHRAWNAGTCCGLPQRNNVDDVGFLTAMVAA IGKQIPIDSARIYATGMSNGAMMALRLGCQTDIFAAIAPVAGALVTDCARPRPTSVLQIHGTADQSVPYD GGPGKAVKINGSPRVDGPSVPSVSATWRAIDACGPPISSTAGVVTTVTADCADGHTVELISVAGAGHQWP GGTRGPLLERLGLPEPSTALDATDTIWQFFAGHRR*
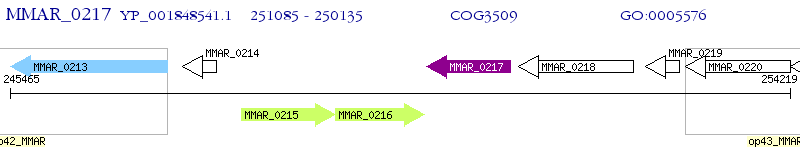
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0217 | - | - | 100% (316) | lipoprotein |
| M. marinum M | MMAR_0227 | lpqP_1 | e-108 | 61.56% (307) | lipoprotein LpqP_1 |
| M. marinum M | MMAR_1000 | lpqP | 3e-71 | 45.52% (290) | lipoprotein, LpqP |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0690 | lpqP | 1e-75 | 48.22% (309) | lipoprotein LpqP |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0671 | lpqP | 1e-75 | 48.22% (309) | lipoprotein LpqP |
| M. leprae Br4923 | MLBr_00715 | lpqC | 8e-38 | 34.77% (279) | putative secreted hydrolase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_5250 | - | 1e-127 | 70.78% (308) | LpqP protein |
| M. smegmatis MC2 155 | MSMEG_3463 | - | 2e-40 | 36.48% (307) | putative esterase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0752 | lpqP | 8e-46 | 47.64% (191) | lipoprotein, LpqP |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0217|M.marinum_M ------MARETARILRMLSAVALGSLVVWTGGCSAGEHAAGAPAPATIPV
MAV_5250|M.avium_104 ------MRFGCARAVSLG---LLPVLVVVLAGCLGGGHALGTPDSQSIPV
Mb0690|M.bovis_AF2122/97 ------MLRRVA---------ILLAAVLAFAGCSG-----GTRLAAGFGN
Rv0671|M.tuberculosis_H37Rv ------MLRRVA---------ILLAAVLAFAGCSG-----GTRLAAGFGN
MUL_0752|M.ulcerans_Agy99 ------MRRHLF---------TLLAVLVALAGCGHR----GAPPPAGFAE
MLBr_00715|M.leprae_Br4923 MNVARWLAS--------------VVLAVCLAGCVGR------QVSASSRD
MSMEG_3463|M.smegmatis_MC2_155 -----MTSS--------------LLGAAMMLGCAAP------WAHAGPLG
**
MMAR_0217|M.marinum_M GRSAQSIQWDGAERTFNLYRPQGLTGAVPLVVMMHGGFGNGAQAERAYHW
MAV_5250|M.avium_104 GPSTHTLQWGGTSRTYHLYRPQGLSEAAPLVVMLHGGFGNGEQAERAYHW
Mb0690|M.bovis_AF2122/97 GNSVHTLDVDGAGRSYRLYKPVGLPSSAPLVVMLHGGFGSAKQAERSYGW
Rv0671|M.tuberculosis_H37Rv GNSVHTLDVDGAGRSYRLYKPVGLPSSAPLVVMLHGGFGSAKQAERSYGW
MUL_0752|M.ulcerans_Agy99 GTSFHTIDVGGVDRTYRLYQPEGLPAGAALVVMLHGGFGNARQAERSDDW
MLBr_00715|M.leprae_Br4923 --QPGTFQFGGLNRTYVLHVPPGS--PVGLVLNLHGGGGTGAGQQGLTDF
MSMEG_3463|M.smegmatis_MC2_155 PIPAAQIDVGGMLRTYQLHLPPGMDKPSGLVVNLHAAGLTGADQAALTHY
:: .* *:: *: * * **: :*.. .. :
MMAR_0217|M.marinum_M NDKADQQHFLVAYPDGLHRAWNA--GTCCGLPQRNNVDDVGFLTAMVAAI
MAV_5250|M.avium_104 DAEADAGHFLVAYPDGLGRAWNA--GTCCGEPAHAGTDDVGFVNAVVGAI
Mb0690|M.bovis_AF2122/97 DELADSEKFLVAYPDGYHRAWNANGGGCCGRPAREGVDDIGFVRAVVADI
Rv0671|M.tuberculosis_H37Rv DELADSEKFLVAYPDGYHRAWNANGGGCCGRPAREGVDDIGFVRAVVADI
MUL_0752|M.ulcerans_Agy99 DELADSEKFVVVYPDGLNRAWNVNGGGCCGRSARDGVDDVAFIGAAVADV
MLBr_00715|M.leprae_Br4923 DAVADLNNLLVAYPDGYDKSWAD--GRGASPADRRHVDDVGFLVELAAKL
MSMEG_3463|M.smegmatis_MC2_155 DSVADAHGFAVVYPDGIDRSWAD--GRGASVPDREGVDDVGFITTLVDRL
: ** : *.**** ::* * .. . : .**:.*: . :
MMAR_0217|M.marinum_M GKQIPIDSARIYATGMSNGAMMALRLGCQ-TDIFAAIAPVAGALVT--DC
MAV_5250|M.avium_104 AAQIPVDRARVYVTGMSNGAMMALRLGCQ-SDTFAAIAPVAGTLLT--DC
Mb0690|M.bovis_AF2122/97 ANNVSIDPARVYVTGMSNGAIMSYTLACN-TSIFAAIGVVSGTQLD--PC
Rv0671|M.tuberculosis_H37Rv ANNVSIDPARVYVTGMSNGAIMSYTLACN-TSIFAAIGVVSGTQLD--PC
MUL_0752|M.ulcerans_Agy99 AHNVGIDANRVYAAGMSNGAMMAYTMACN-TGIFAAIGPVSGTQLD--PC
MLBr_00715|M.leprae_Br4923 QSDFGVAPGHVFVTGMSNGGFMSNKLACDRADMFAAIAPVAGTLGVDVAC
MSMEG_3463|M.smegmatis_MC2_155 VTDFGIPRDRVYATGLSAGAFMANRLACERADLFAAIAPVAGTLGSNVGC
:. : :::.:*:* *.:*: :.*: :. ****. *:*: *
MMAR_0217|M.marinum_M ARPRPTSVLQIHGTADQSVPYDGGPGKAVKINGSPRVDGPSVPSVSATWR
MAV_5250|M.avium_104 SAARPASVLQIHGTADDRVPYAGGPGKAFALNGSPRVDGPSVESVNATWR
Mb0690|M.bovis_AF2122/97 QSPRPVSVIHIHGTADPLVRYHGGPGAGFAR-----IDGPPVPDLNAFWR
Rv0671|M.tuberculosis_H37Rv QSPRPVSVIHIHGTADPLVRYHGGPGAGFAR-----IDGPPVPDLNAFWR
MUL_0752|M.ulcerans_Agy99 RSPHPVSGCTFT---APEIRWFATRGA----------PGPAPP-------
MLBr_00715|M.leprae_Br4923 HPSQPVSVMDAHGTDDPLVPFNGGDVHGRGG----VSHVVSVTNMVEKWR
MSMEG_3463|M.smegmatis_MC2_155 SPSQPVSVLAMYGTADPIVPFGGGVMNGRGG----TSTVLSATELVDRWR
.:*.* : : . .
MMAR_0217|M.marinum_M AIDACGPPISSTAGVVTTV--------TADCADGHTVELISVAGAGHQWP
MAV_5250|M.avium_104 AIDACGPPSSTTAGDVTTQ--------TAGCADGRTVELISVAGAGHQWP
Mb0690|M.bovis_AF2122/97 EVNRCGALDTTTEGPVTTS--------GATCADNRRVVLLTVDDAGHRWP
Rv0671|M.tuberculosis_H37Rv EVNRCGALDTTTEGPVTTS--------GATCADNRRVVLLTVDDAGHRWP
MUL_0752|M.ulcerans_Agy99 --------------PILR---------GRRCRT-----------------
MLBr_00715|M.leprae_Br4923 TADGCQGDPSQQELPDVGDGTFVRRFDYTVCANSAEVVFYKINKGGHTWP
MSMEG_3463|M.smegmatis_MC2_155 GLDGCADDPAPETLPAVGDGTSTERLTYAPCAAGTAVVFMRVDNGGHTWP
*
MMAR_0217|M.marinum_M GGTRGPLLERLG-LPEPSTALDATDTIWQFFAGHRR-
MAV_5250|M.avium_104 GGAPSPLAEKVAGIPAPSTALDATDTIWQFFARNHR-
Mb0690|M.bovis_AF2122/97 S--------------------FATQTLWRFFAAHFR-
Rv0671|M.tuberculosis_H37Rv S--------------------FATQTLWRFFAAHFR-
MUL_0752|M.ulcerans_Agy99 -------------------------------------
MLBr_00715|M.leprae_Br4923 GGKQYLPKVVIG---STTCVFDASEVIAQFFLTHARS
MSMEG_3463|M.smegmatis_MC2_155 GAPEVLPAQQVG---LAIRGFDASVLSMDFFEAHGR-