For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SETYEQLHGGRAVASSFVPRHIGPDPAAIAHMLATVGYPSLDDLIDAAVPARIRNTAPMALPVARTEQQV LTDLASMAERNTIRVSMIGQGYSPCVTPEVLRRNLIESPAWYTAYTPYQPEISQGRLEALLTFQTMIEDL TGLPLAGASLLDEATAVVEAMLLMRRSNKSPSARLLVDSDCFTQTLSVLRTRAAAVGIDIELVDIEKKLP EGDFFGIVLQAPGASGAVRHLGRLIATAKARGAMTTVAADLLSLTLLTPPGELGADIAVGSAQRFGVPLF FGGPHAGYMAVRSGLERALPGRLVGVSKDVNGAPAYRLALQTREQHIRRDKATSNICTAQALLANVAAMY AVYHGPEGLRDIASRVHAHAVSIAASLRHHGYTVAHDQFFDTVLVEVPGRAGELIARAESLGINLRRAGS DHVGISCDETTTTEVITLVLKAFDVERIAPSRSPLGSELLRTTPYLQHGVFHEHRSETAMMRFLRKLSDR DLALDRTMIPLGSCTMKLNSAVEMAPISWPEFAAIHPHAPIAQTTGYRELIADLERWLAEITGYDRVSLQ PNAGSQGELAGLLAIHGYHASRGEQQRRICLTPQSAHGTNAASAVLAGMQVVVVATAADGSIDLADLRAK ISTHSAELAAIMLTYPSTHGVYEPDVTTVCDLVHKAGGQVYIDGANLNALVGIARPGHFGGDVSHLNLHK TFCIPHGGGGPGVGPVAVRAHLAPFLPDDPHQDTSDSSVAAANYGSAGILPITWAYIALMGPDGLATATK SAVLSANYLAARLNDHYPVLYTGPHGLVAHECILDLRELTRRSGVTAEDVAKRLIDYGFHAPTLAFPVSG TLMVEPTESEDLAELDRFVDAMVAIRGEIDLVMWGEWPLEASPLRQAPHTAEMVCADTWDLPYPRSVAAF PAPWLVTDKYWPPVRRIDGVHGDRNLVCSCPSPAAFESTLPPKSASLQVL*
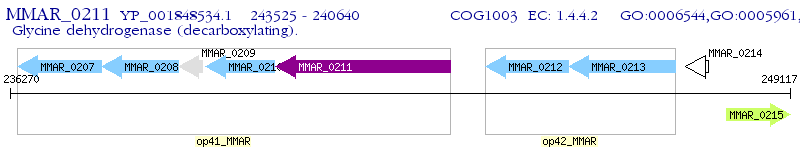
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0211 | gcvB_1 | - | 100% (961) | glycine dehydrogenase GcvB_1 |
| M. marinum M | MMAR_2708 | gcvB | 0.0 | 62.21% (942) | glycine dehydrogenase GcvB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1863 | gcvB | 0.0 | 62.53% (942) | glycine dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3398 | - | 0.0 | 62.21% (942) | glycine dehydrogenase |
| M. tuberculosis H37Rv | Rv1832 | gcvB | 0.0 | 62.53% (942) | glycine dehydrogenase |
| M. leprae Br4923 | MLBr_02072 | gcvB | 0.0 | 61.15% (942) | glycine dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2407 | - | 0.0 | 63.15% (947) | glycine dehydrogenase |
| M. avium 104 | MAV_2884 | gcvP | 0.0 | 63.63% (943) | glycine dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3642 | gcvP | 0.0 | 63.30% (940) | glycine dehydrogenase |
| M. thermoresistible (build 8) | TH_0290 | gcvB | 0.0 | 64.63% (557) | Probable glycine dehydrogenase gcvB (Glycine decarboxylase) |
| M. ulcerans Agy99 | MUL_3050 | gcvB | 0.0 | 62.00% (942) | glycine dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3131 | - | 0.0 | 62.70% (941) | glycine dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3398|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3131|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3642|M.smegmatis_MC2_155 --------------------------------------------------
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 --------------------------------------------------
Rv1832|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3050|M.ulcerans_Agy99 --------------------------------------------------
MAV_2884|M.avium_104 --------------------------------------------------
MLBr_02072|M.leprae_Br4923 --------------------------------------------------
MAB_2407|M.abscessus_ATCC_1997 MTASHDRRRSNTSPSISQAPEPPAAPMPLKSGDPVTEPDRPPTGKGFYQE
MMAR_0211|M.marinum_M --------------------------------------------------
Mflv_3398|M.gilvum_PYR-GCK ---------------------------MPDQTPAPSVLRFVDRHIGPDDQ
Mvan_3131|M.vanbaalenii_PYR-1 ---------------------------MSDQFRTPS---FVDRHIGPDAH
MSMEG_3642|M.smegmatis_MC2_155 ---------------------------MSDQPKARNP-QFVDRHIGPDES
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 ---------------------------MSDH------STFADRHIGLDSQ
Rv1832|M.tuberculosis_H37Rv ---------------------------MSDH------STFADRHIGLDSQ
MUL_3050|M.ulcerans_Agy99 ---------------------------MSDHS--MFESTFADRHIGLDNQ
MAV_2884|M.avium_104 ---------------------------MPDH------TTFAARHIGPDPQ
MLBr_02072|M.leprae_Br4923 -------------------------MSVPNSS--NKQTCFTARHIGPNSE
MAB_2407|M.abscessus_ATCC_1997 ILRVERPNLSGARDGLMTEGGETLSRAVSRDSASGSAMSFANRHIGPTTA
MMAR_0211|M.marinum_M ----------------------MSETYEQLHGGRAVASSFVPRHIGPDPA
Mflv_3398|M.gilvum_PYR-GCK AVETLLNTIGVPSLDELAAKALPDVILDRLSTDGVAPGLEHLPAAATEHE
Mvan_3131|M.vanbaalenii_PYR-1 AVATLLGTIGVSSLDELAAKALPAGILDPLTGAGTAPGLEHLPPAATEHE
MSMEG_3642|M.smegmatis_MC2_155 AVATMLDVIGVSSLDELAAKALPTGILDALDAAGRAPGLAELPPAATEEE
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 AVATMLAVIGVDSLDDLAVKAVPAGILDTLTDTGAAPGLDSLPPAASEAE
Rv1832|M.tuberculosis_H37Rv AVATMLAVIGVDSLDDLAVKAVPAGILDTLTDTGAAPGLDSLPPAASEAE
MUL_3050|M.ulcerans_Agy99 AIATMLAVIGVDSLDDLAAKAVPTRIFDTPTDSGLAPGLDQLPPAASEAE
MAV_2884|M.avium_104 AVAAMLDVIGVGSLDELAAKAVPAGIRDRLSADGIAPGLDRLPPPASETE
MLBr_02072|M.leprae_Br4923 DVATMLAVIGVESLDDLAAKAVPSDILDNVTDTGVAPGLDRLPPPATESE
MAB_2407|M.abscessus_ATCC_1997 DIDTMLATIGVASLDELAEKAVPASILDALR-DGLASGLDALPVAASEHE
MMAR_0211|M.marinum_M AIAHMLATVGYPSLDDLIDAAVPARIR------NTAP--MALPVARTEQQ
Mflv_3398|M.gilvum_PYR-GCK ALAELRALAQSNTVAVSMIGQGYYDTLTPPVLRRNIIENPAWYTAYTPYQ
Mvan_3131|M.vanbaalenii_PYR-1 ALAELRALAESNTVAVSMIGQGYYDTLTPPVLRRNILENPAWYTAYTPYQ
MSMEG_3642|M.smegmatis_MC2_155 ALAELRALADTNTVAVSMIGQGYFDTLTPAVLRRNILENPAWYTAYTPYQ
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 ALAELRALADANTVAVSMIGQGYYDTHTPPVLLRNIIENPAWYTAYTPYQ
Rv1832|M.tuberculosis_H37Rv ALAELRALADANTVAVSMIGQGYYDTHTPPVLLRNIIENPAWYTAYTPYQ
MUL_3050|M.ulcerans_Agy99 ALAELRALADTNTTAVSMIGQGYYDTFTPQVLLRNIMENPAWYTAYTPYQ
MAV_2884|M.avium_104 ALAELRGLAEANTVAVSMIGQGYYDTLTPPVLLRNILENPAWYTAYTPYQ
MLBr_02072|M.leprae_Br4923 TLAELGALARANTVAVSMIGQGYYDTLTPAVLSRNILENPAWYTPYTPYQ
MAB_2407|M.abscessus_ATCC_1997 ALDALRKLASQNTVAVSMIGQGYFDTLTPPVLRRHILENPAWYTAYTPYQ
MMAR_0211|M.marinum_M VLTDLASMAERNTIRVSMIGQGYSPCVTPEVLRRNLIESPAWYTAYTPYQ
Mflv_3398|M.gilvum_PYR-GCK PEISQGRLEALLNFQTMVTDLTGLEVANASMLDEGTAAAEAMTLMHRAVR
Mvan_3131|M.vanbaalenii_PYR-1 PEISQGRLEALLNFQTMVSDLTGLEVANASMLDEGTAAAEAMTLMHRAVR
MSMEG_3642|M.smegmatis_MC2_155 PEISQGRLEALLNFQTMVCDLTGLEVANASMLDEGTAAAEAMTLMHRATR
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 PEISQGRLEALLNFQTLVTDLTGLEIANASMLDEGTAAAEAMTLMHRAAR
Rv1832|M.tuberculosis_H37Rv PEISQGRLEALLNFQTLVTDLTGLEIANASMLDEGTAAAEAMTLMHRAAR
MUL_3050|M.ulcerans_Agy99 PEISQGRLEALLNFQTMVTDLTGLEIANASMLDEGTAAAEAMTLMHRATR
MAV_2884|M.avium_104 PEISQGRLEALLNFQTMVADLTGLEIANASMLDEGTAAAEAMTLMHRASR
MLBr_02072|M.leprae_Br4923 PEISQGRLEALLNFQTLVSDLTGLEIANASMLDEGTAAAEAMTLMYRAAR
MAB_2407|M.abscessus_ATCC_1997 PEISQGRLEALLNFQTMVAELTGLDIANASMLDEGTAAAEAMTLMHRAVK
MMAR_0211|M.marinum_M PEISQGRLEALLTFQTMIEDLTGLPLAGASLLDEATAVVEAMLLMRRSNK
Mflv_3398|M.gilvum_PYR-GCK GPATRLAVDADVYPQTAAILATRAEPLGIEIVTADLRQGLPDGDFFGVIV
Mvan_3131|M.vanbaalenii_PYR-1 GPSNRLVVDSDVYAQTAAVLATRAEPLGIEIVTADLRHGLPDGDFFGVIV
MSMEG_3642|M.smegmatis_MC2_155 SKVNRLLVDTDVYAQTAAVLATRARPLGIEIVTADLSKGLPDGEFFGVVV
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 GPVKRVVVDADVFTQTAAVLATRAKPLGIEIVTADLRAGLPDGEFFGAIA
Rv1832|M.tuberculosis_H37Rv GPVKRVVVDADVFTQTAAVLATRAKPLGIEIVTADLRAGLPDGEFFGVIA
MUL_3050|M.ulcerans_Agy99 SSTKHLAVDADLFTQTAAVLATRAKPLGIEIVTTDLRDGLPDGEFFGVIA
MAV_2884|M.avium_104 GKSNRLAVDVDVFAQTAAIVATRARPLGIEIVTADLRDGLPDGDFFGVIA
MLBr_02072|M.leprae_Br4923 STASRVVVDVDVFAQTVAVFATRAKPLGIDIVVADLREGLPDGEFFGVIT
MAB_2407|M.abscessus_ATCC_1997 SASNRLVVDADLYPQTAAVIATRAEPLGIDVVTADLAAGLPEGDFFGVIV
MMAR_0211|M.marinum_M SPSARLLVDSDCFTQTLSVLRTRAAAVGIDIELVDIEKKLPEGDFFGIVL
Mflv_3398|M.gilvum_PYR-GCK QLPGASGVVHDWSALVEQAHERGALVAVGADLLAATMITPPGEIGADVAF
Mvan_3131|M.vanbaalenii_PYR-1 QLPGAGGAITDWSELVTQAHDRGALIAVGADLLALTLIAPPGEIGADVAF
MSMEG_3642|M.smegmatis_MC2_155 QLPGASGAVVDWAPLIEQAHERKALVAVGADLLALTLITPPGEIGADVAF
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 QLPGASGRITDWSALVQQAHDRGALVAVGADLLALTLIAPPGEIGADVAF
Rv1832|M.tuberculosis_H37Rv QLPGASGRITDWSALVQQAHDRGALVAVGADLLALTLIAPPGEIGADVAF
MUL_3050|M.ulcerans_Agy99 QLPGASGRITDWTALIQQAHDRGALVSIGADLLALTLLTPPGEIGADIAF
MAV_2884|M.avium_104 QLPGASGAITDWAALVAQAHERGALVALGADLLALTLITPPGEIGADVAF
MLBr_02072|M.leprae_Br4923 QLPGASGRITDWTALIAQAHSRGALVAVGADLLALTLITPPGEIGADVAF
MAB_2407|M.abscessus_ATCC_1997 QLPGASGVVRDWTTLISAAHERGALVAVGADLLALTLVAPPGDIGADVAF
MMAR_0211|M.marinum_M QAPGASGAVRHLGRLIATAKARGAMTTVAADLLSLTLLTPPGELGADIAV
Mflv_3398|M.gilvum_PYR-GCK GTTQRFGVPMGFGGPHAGYLAVHSKHARQLPGRLVGVSVDADGSRAYRLA
Mvan_3131|M.vanbaalenii_PYR-1 GTTQRFGVPMGFGGPHAGYLAVHSKHARQLPGRLVGVSVDADGAAAFRLA
MSMEG_3642|M.smegmatis_MC2_155 GTTQRFGVPMGFGGPHAGYLAVHANHARQLPGRLVGVSVDSDGSPAYRLA
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 GTTQRFGVPMGFGGPHAGYLAVHAKHARQLPGRLVGVSVDSDGTPAYRLA
Rv1832|M.tuberculosis_H37Rv GTTQRFGVPMGFGGPHAGYLAVHAKHARQLPGRLVGVSVDSDGTPAYRLA
MUL_3050|M.ulcerans_Agy99 GTTQRFGVPMGFGGPHAGYLSVHAKHARQLPGRLVGVSVDNDGNPAYRLA
MAV_2884|M.avium_104 GTTQRFGVPMGFGGPHAGYLAVHANHARQLPGRLVGVSLDADGSPAYRLA
MLBr_02072|M.leprae_Br4923 GTTQRFGVPMGFGGPHAGYLALHTKHARQLPGRLVGVSVDSDGTPAYRLA
MAB_2407|M.abscessus_ATCC_1997 GTTQRFGVPMGFGGPHAGYLAVHTAHARQLPGRLVGVSVDADGSPAYRLS
MMAR_0211|M.marinum_M GSAQRFGVPLFFGGPHAGYMAVRSGLERALPGRLVGVSKDVNGAPAYRLA
Mflv_3398|M.gilvum_PYR-GCK LQTREQHIRRDKATSNICTAQVLLAVLAAMYASYHGPDGLRGIAQRVHGH
Mvan_3131|M.vanbaalenii_PYR-1 LQTREQHIRRDKATSNICTAQVLLAVMAAMYASYHGADGLRAIALRVHAQ
MSMEG_3642|M.smegmatis_MC2_155 LQTREQHIRRDKATSNICTAQVLLAVIAAMYASYHGAPGLTAIARRVHAH
TH_0290|M.thermoresistible__bu --------------------------------------------------
Mb1863|M.bovis_AF2122/97 LQTREQHIRRDKATSNICTAQVLLAVLAAMYASYHGAGGLTAIARRVHAH
Rv1832|M.tuberculosis_H37Rv LQTREQHIRRDKATSNICTAQVLLAVLAAMYASYHGAGGLTAIARRVHAH
MUL_3050|M.ulcerans_Agy99 LQTREQHIRRDKATSNICTAQVLLAVMAAMYASYHGADGLTGIARRVHGH
MAV_2884|M.avium_104 LQTREQHIRRDKATSNICTAQVLLAVMAAMYASYHGAEGLTAIARRVHGH
MLBr_02072|M.leprae_Br4923 LQTREQHIRRDKATSNICTAQVLLAVMAAMYASYHGAEGLTGIARRVHAQ
MAB_2407|M.abscessus_ATCC_1997 LQTREQHIRRDKATSNICTAQVLLAVMAAMYASYHGPEGLRAIAARIHRN
MMAR_0211|M.marinum_M LQTREQHIRRDKATSNICTAQALLANVAAMYAVYHGPEGLRDIASRVHAH
Mflv_3398|M.gilvum_PYR-GCK ARALAAGLADAGVEVVHDSFFDTVLAHVPGRADEVRAAAKERGINVWAVD
Mvan_3131|M.vanbaalenii_PYR-1 ASALAAGLSGAGIEVVHPSFFDTVLARVPGRASEVRAAAKERGINVWLVD
MSMEG_3642|M.smegmatis_MC2_155 ADAIASALGDA---VVHDKFFDTVLVRVPGRADEVVAAAKAKNINLWRVD
TH_0290|M.thermoresistible__bu -----------------------VLARVPGRAEQIRAAAKERGINLWLVD
Mb1863|M.bovis_AF2122/97 AEAIAG---ALGDALVHDKYFDTVLARVPGRADEVLARAKANGINLWRVD
Rv1832|M.tuberculosis_H37Rv AEAIAG---ALGDALVHDKYFDTVLARVPGRADEVLARAKANGINLWRVD
MUL_3050|M.ulcerans_Agy99 AETIAG---ALGDALVHDTFFDTVLARVPDRADEVVAAAKASGVNLWRVD
MAV_2884|M.avium_104 AEAIAA---ALGTAVVHDRYFDTVLARVPGRAHEVIAAAKARGINLWRVD
MLBr_02072|M.leprae_Br4923 ARALAAGLSAAGVEVVHQAFFDTVLARVPGRTVQIQGAAKERGINVWLVD
MAB_2407|M.abscessus_ATCC_1997 AQLLASGLTRGGHTVVHDRFFDTVLVQVPGRAAQIQAAAKAEGINIWSPD
MMAR_0211|M.marinum_M AVSIAASLRHHGYTVAHDQFFDTVLVEVPGRAGELIARAESLGINLRRAG
**..**.*: :: . *: .:*: .
Mflv_3398|M.gilvum_PYR-GCK ADHVSVACDEATTAEHVADVLAAFG--AAP-SGADFAGPAVATRTSEFLT
Mvan_3131|M.vanbaalenii_PYR-1 DDHISVSCDEATTDGHVADVLAAFS--AEPTVGGDVTAASIVTRISEFLT
MSMEG_3642|M.smegmatis_MC2_155 DDHVSVSCDEATTDAHIDAVIKAFG--ATR---TDGAGTDIATRTSEFLT
TH_0290|M.thermoresistible__bu ADHVSVSCDEATRPEHVTEVLAAFG--ATP-VDEPFTGPDIATRTTPFLT
Mb1863|M.bovis_AF2122/97 ADHVSVACDEATTDTHVAVVLDAFG--VAA-AAP--AHADIATRTSEFLT
Rv1832|M.tuberculosis_H37Rv ADHVSVACDEATTDTHVAVVLDAFG--VAA-AAP--AHTDIATRTSEFLT
MUL_3050|M.ulcerans_Agy99 ADHVSVACDEVTTAAHVTAVLDAFG--VSA-TKA--ARAEIATRTSEFLT
MAV_2884|M.avium_104 DDHVSVACDEATTDEHVAAVLEAFG--VAP-AEP--VASEIATRTSEFLT
MLBr_02072|M.leprae_Br4923 GDHVSVACDEATTDEHITAVLAAFA--ATP-ARASFAGPDIATRTSAFLT
MAB_2407|M.abscessus_ATCC_1997 GDHVSIACDEATTVPHLVAVLRAFGSEIGQDTAGAPGASDIATRGSDYLT
MMAR_0211|M.marinum_M SDHVGISCDETTTTEVITLVLKAFD--VERIAPSRSPLGSELLRTTPYLQ
**:.::***.* : *: ** * : :*
Mflv_3398|M.gilvum_PYR-GCK HPAFSDYRTETSMMRYLRSLADKDIALDRSMIPLGSCTMKLNAAAEMEAI
Mvan_3131|M.vanbaalenii_PYR-1 HPAFTRYRTETEMMRYLRALADKDIALDRSMIPLGSCTMKLNAAAEMEPI
MSMEG_3642|M.smegmatis_MC2_155 HPAFHRYRTETEMMRYLRSLADKDIALDRSMIPLGSCTMKLNAAAEMEPI
TH_0290|M.thermoresistible__bu HPAFTRYRTETEMMRYLRSLADKDIALDRSMIPLGSCTMKLNAAAEMEPI
Mb1863|M.bovis_AF2122/97 HPAFTQYRTETSMMRYLRALADKDIALDRSMIPLGSCTMKLNAAAEMESI
Rv1832|M.tuberculosis_H37Rv HPAFTQYRTETSMMRYLRALADKDIALDRSMIPLGSCTMKLNAAAEMESI
MUL_3050|M.ulcerans_Agy99 HPAFTNYRTETSMMRYLRELADKDIALDRSMIPLGSCTMKLNAATEMESI
MAV_2884|M.avium_104 HPAFTQYRTETAMMRYLRTLADKDIALDRSMIPLGSCTMKLNAAAEMEPI
MLBr_02072|M.leprae_Br4923 HPTFTKYRTETSMMRYLRALADKDIALDRSMIPLGSCTMKLNAAAEMESI
MAB_2407|M.abscessus_ATCC_1997 HPAFNRYHSETEMMRYLRALSDKDIALDRSMIPLGSCTMKLNAAAEMEPI
MMAR_0211|M.marinum_M HGVFHEHRSETAMMRFLRKLSDRDLALDRTMIPLGSCTMKLNSAVEMAPI
* .* :::** ***:** *:*:*:****:************:*.** .*
Mflv_3398|M.gilvum_PYR-GCK TWAEFGRQHPFAPASDTPGLRRLIADLQSWLTGITGYDEISLQPNAGSQG
Mvan_3131|M.vanbaalenii_PYR-1 TWPEFARQHPFAPASDTPGIRRLIADLQGWLTGITGYDEISLQPNAGSQG
MSMEG_3642|M.smegmatis_MC2_155 TWTEFSRQHPFAPASDTPGLRKLIADLQSWLTAITGYDEVSLQPNAGSQG
TH_0290|M.thermoresistible__bu TWPEFTRLHPFAPKEDTPGIRRLIADLEQWLAHITGYAAVSLQPNAGSQG
Mb1863|M.bovis_AF2122/97 TWPEFGRQHPFAPASDTAGLRQLVADLQSWLVLITGYDAVSLQPNAGSQG
Rv1832|M.tuberculosis_H37Rv TWPEFGRQHPFAPASDTAGLRQLVADLQSWLVLITGYDAVSLQPNAGSQG
MUL_3050|M.ulcerans_Agy99 TWPELGRQHPFAPASDTPGLRRLITELQGWLAAITGYDEVSLQPNAGSQG
MAV_2884|M.avium_104 TWPEFARQHPFAPASDTPGLRRLIGDLENWLVAITGYDAVSLQPNAGSQG
MLBr_02072|M.leprae_Br4923 TWQEFTRQHPFAPVSDTPGLRRLISDLESWLVQITGYDAVSLQPNAGSQG
MAB_2407|M.abscessus_ATCC_1997 TWPQFAALHPFAPAGDSRGLRTLIADLEGWLADITGYDKVSLQPNAGSQG
MMAR_0211|M.marinum_M SWPEFAAIHPHAPIAQTTGYRELIADLERWLAEITGYDRVSLQPNAGSQG
:* :: **.** :: * * *: :*: **. **** :**********
Mflv_3398|M.gilvum_PYR-GCK EYAGLLAIQAYHHARGDSGRTVCLIPSSAHGTNAASAAMVGMKVVVVACR
Mvan_3131|M.vanbaalenii_PYR-1 EYAGLLAIRAYHAARGDTDRTVCLIPASAHGTNAASAAMVGMQVVVVACR
MSMEG_3642|M.smegmatis_MC2_155 EYAGLLAIKAYHEARGDADRDICLIPSSAHGTNAASAALAGMRVVVVACR
TH_0290|M.thermoresistible__bu EYAGLLAIQAYHAERGEPQRDVCLIPSSAHGTNAASAALAGMRVVVVGCR
Mb1863|M.bovis_AF2122/97 EYAGLLAIHEYHASRGEPHRDICLIPSSAHGTNAASAALAGMRVVVVDCH
Rv1832|M.tuberculosis_H37Rv EYAGLLAIHEYHASRGEPHRDICLIPSSAHGTNAASAALAGMRVVVVDCH
MUL_3050|M.ulcerans_Agy99 EYSGLLAIHEYHASRGEPHRDICLIPSSAHGTNAASAALAGMRVVVVGCH
MAV_2884|M.avium_104 EYAGLLAIHDYHASRGEPHRDICLIPSSAHGTNAASAALAGMRVVVVGCH
MLBr_02072|M.leprae_Br4923 EYAGLLAIHDYHVSRGEPHRNVCLIPSSAHGTNAASAALVGMRVVVVGCH
MAB_2407|M.abscessus_ATCC_1997 EYAGLLAIRQYHIDRGDSGRDVCLIPSSAHGTNAASAALAGMRVVVVACR
MMAR_0211|M.marinum_M ELAGLLAIHGYHASRGEQQRRICLTPQSAHGTNAASAVLAGMQVVVVATA
* :*****: ** **: * :** * **********.:.**:****
Mflv_3398|M.gilvum_PYR-GCK ANGDVDLDDLRAKVTEHADRLSALMITYPSTHGVYEHDIADICAAVHDAG
Mvan_3131|M.vanbaalenii_PYR-1 ANGDVDLDDLRAKVTEHAERLAALMITYPSTHGVYEHDIAEICAAVHDAG
MSMEG_3642|M.smegmatis_MC2_155 ENGDVDLDDLRTKVKEHADRIAALMITYPSTHGVYEHDVADICAAVHDVG
TH_0290|M.thermoresistible__bu PNGDVDLDDLRVKIAEHSDRLAALMITYPSTHGVYEHDVAEICAAVHDAG
Mb1863|M.bovis_AF2122/97 DNGDVDLDDLRAKVGEHAERLSALMITYPSTHGVYEHDIAEICAAVHDAG
Rv1832|M.tuberculosis_H37Rv DNGDVDLDDLRAKVGEHAERLSALMITYPSTHGVYEHDIAEICAAVHDAG
MUL_3050|M.ulcerans_Agy99 DNGDVDLDDLRAKVGEHADRLAALMITYPSTHGVYEHDIADICAAVHDAG
MAV_2884|M.avium_104 DNGDVDLDDLRAKVTDHRDRLSTLMITYPSTHGVYEHDIAEICAAVHDAG
MLBr_02072|M.leprae_Br4923 DNGDVDLDDLRIKLSEHANRLSVLMITYPSTHGVYEHDIAEICAAVHDAG
MAB_2407|M.abscessus_ATCC_1997 ENGDVDLDDLRAKIAANASALSAIMITYPSTHGVYEHDIADICAAVHDAG
MMAR_0211|M.marinum_M ADGSIDLADLRAKISTHSAELAAIMLTYPSTHGVYEPDVTTVCDLVHKAG
:*.:** *** *: : ::.:*:********** *:: :* **..*
Mflv_3398|M.gilvum_PYR-GCK GQVYVDGANLNALVGLARPGRFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
Mvan_3131|M.vanbaalenii_PYR-1 GQVYVDGANLNALVGLARPGRFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MSMEG_3642|M.smegmatis_MC2_155 GQVYVDGANLNALVGLARPGRFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
TH_0290|M.thermoresistible__bu GQVYVDGANLNALVGLARPGRFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
Mb1863|M.bovis_AF2122/97 GQVYVDGANLNALVGLARPGKFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
Rv1832|M.tuberculosis_H37Rv GQVYVDGANLNALVGLARPGKFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MUL_3050|M.ulcerans_Agy99 GQVYVDGANLNALVGLARPGKFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MAV_2884|M.avium_104 GQVYVDGANLNALVGLARPGKFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MLBr_02072|M.leprae_Br4923 GQVYVDGANLNALVGLARPGKFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MAB_2407|M.abscessus_ATCC_1997 GQVYVDGANLNALVGLARPGHFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
MMAR_0211|M.marinum_M GQVYIDGANLNALVGIARPGHFGGDVSHLNLHKTFCIPHGGGGPGVGPVA
****:**********:****:*****************************
Mflv_3398|M.gilvum_PYR-GCK VRAHLAPYLPGHPLAAELSDDHTVSAAPYGSASILPITWAYIRMMGAAGL
Mvan_3131|M.vanbaalenii_PYR-1 VRSHLAPYLPGHPLADELSDEHTVSAAPYGSASILPITWAYIRMMGAQGL
MSMEG_3642|M.smegmatis_MC2_155 VRAHLAPYLPGHPLADELSDDHTVSAAPYGSASILPITWMYIRMMGADGL
TH_0290|M.thermoresistible__bu VGVHLAKYLPGHPLADELGDTLTVSAAPYGSASILPITWMYIRMMGAQGL
Mb1863|M.bovis_AF2122/97 VRAHLAPFLPGHPFAPELPKGYPVSSAPYGSASILPITWAYIRMMGAEGL
Rv1832|M.tuberculosis_H37Rv VRAHLAPFLPGHPFAPELPKGYPVSSAPYGSASILPITWAYIRMMGAEGL
MUL_3050|M.ulcerans_Agy99 VRSHLAPFLPGHPFAPGLPQGDPVSAAPYGSASILPITWAYIRMMGADGL
MAV_2884|M.avium_104 VRSHLAPFLPGHPHAPELPQGHPVSSAPYGSASILPISWAYIRMMGADGL
MLBr_02072|M.leprae_Br4923 VRSHLVSFLPGHPFAPELPQGQPVSSAPYGSASLLPITWAYIRMMGADGL
MAB_2407|M.abscessus_ATCC_1997 VRSHLAQYLPGHPYAEELPAGAPVSSAPYGSASILPITWAYIRMMGPDGL
MMAR_0211|M.marinum_M VRAHLAPFLPDDPHQDTS--DSSVAAANYGSAGILPITWAYIALMGPDGL
* **. :**..* .*::* ****.:***:* ** :**. **
Mflv_3398|M.gilvum_PYR-GCK RSATLVAIASANYIARRLDEYYPVLYTGENGMVAHECILDLRGITKATGV
Mvan_3131|M.vanbaalenii_PYR-1 RSASLVAIASANYIARRLDEYYPVLYTGENGMVAHECILDLRAITKATGV
MSMEG_3642|M.smegmatis_MC2_155 RAATLTAIASANYVARRLDEYFPVLYTGENGMVAHECILDLRGITKATGV
TH_0290|M.thermoresistible__bu RTATLTAIASANYIARRLDEYFPVLYTGENGMVAHECILDLRPLTKATGV
Mb1863|M.bovis_AF2122/97 RAASLTAITSANYIARRLDEYYPVLYTGENGMVAHECILDLRGITKLTGI
Rv1832|M.tuberculosis_H37Rv RAASLTAITSANYIARRLDEYYPVLYTGENGMVAHECILDLRGITKLTGI
MUL_3050|M.ulcerans_Agy99 RAASLTAIASANYIARRLDEYFPVLYTGENGMVAHECILDLRGIPKATGV
MAV_2884|M.avium_104 RAASLTAITSANYIARRLDEYFPVLYTGENGMVAHECILDLRPITKATGV
MLBr_02072|M.leprae_Br4923 RTASLTAIASANYIARRLDKYFPVLYTGENGMVAHECILDLRPITKSVGV
MAB_2407|M.abscessus_ATCC_1997 RAASLTAIASANYLARRLDEYYPVLYTGDNGMVAHECILDLREITKNTGV
MMAR_0211|M.marinum_M ATATKSAVLSANYLAARLNDHYPVLYTGPHGLVAHECILDLRELTRRSGV
:*: *: ****:* **:.::****** :*:********** :.: *:
Mflv_3398|M.gilvum_PYR-GCK TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLSEIDAFCDAMIAI
Mvan_3131|M.vanbaalenii_PYR-1 TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLAEVDAFCEAMIAI
MSMEG_3642|M.smegmatis_MC2_155 TVDDVAKRLADFGFHAPTMSFPVSGTLMVEPTESESLAEVDAFCDAMIAI
TH_0290|M.thermoresistible__bu TVDDVAKRLADFGFHAPTMSFPVAGTLMVEPTESESLAEIDAFCEAMIAI
Mb1863|M.bovis_AF2122/97 TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLAEVDAFCEAMIGI
Rv1832|M.tuberculosis_H37Rv TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLAEVDAFCEAMIGI
MUL_3050|M.ulcerans_Agy99 TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLAEVDAFCDAMISI
MAV_2884|M.avium_104 TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESETLTEVDAFCDAMIAI
MLBr_02072|M.leprae_Br4923 TVDDVAKRLADYGFHAPTMSFPVPGTLMVEPTESESLAEIDAFCEAMIAI
MAB_2407|M.abscessus_ATCC_1997 TVDDVAKRLADYGFHAPTMSFPVAGTLMVEPTESESLAEVDAFCEAMIAI
MMAR_0211|M.marinum_M TAEDVAKRLIDYGFHAPTLAFPVSGTLMVEPTESEDLAELDRFVDAMVAI
*.:****** *:******::***.*********** *:*:* * :**:.*
Mflv_3398|M.gilvum_PYR-GCK RAEIDRVGSGEWPVDDNPLRGAPHTAESLLVEEWTHPYTREQAAYPLGKG
Mvan_3131|M.vanbaalenii_PYR-1 RGEIDKVGSGMWSVDDNPLRGAPHTAESLLVEDWHHPYTREQAAYPLGKG
MSMEG_3642|M.smegmatis_MC2_155 RGEIDRVGSGEWPVDDNPLRGAPHTADCLLVADWNRPYTREEAAYPLGKG
TH_0290|M.thermoresistible__bu RREIDRVGSGEWPVDDNPLRNAPHTAECLIAGEWNHPYTREQAAYPLGTG
Mb1863|M.bovis_AF2122/97 RAEIDKVGAGEWPVDDNPLRGAPHTAQCLLASDWDHPYTREQAAYPLGTA
Rv1832|M.tuberculosis_H37Rv RAEIDKVGAGEWPVDDNPLRGAPHTAQCLLASDWDHPYTREQAAYPLGTA
MUL_3050|M.ulcerans_Agy99 RAEIDRVGSGQWPVDDNPLRGAPHTAQCLLVGEWDHPYTREEAAYPLGTA
MAV_2884|M.avium_104 RGEIDRVGAGEWPVEDNPLRGAPHTAECLVTTDWDHPYSREQAAYPLGKD
MLBr_02072|M.leprae_Br4923 RGEIARVGAGEWSVEDNPLRGAPHTAECLLASDWDHPYTREEAAYPLGKA
MAB_2407|M.abscessus_ATCC_1997 KSEIDKVGSGVWPAQDNPLRGAPHTAESLIADEWHHPYTRAEAAYPLGRG
MMAR_0211|M.marinum_M RGEIDLVMWGEWPLEASPLRQAPHTAEMVCADTWDLPYPRSVAAFPAPWL
: ** * * *. : .*** *****: : . * **.* **:*
Mflv_3398|M.gilvum_PYR-GCK FRPKVWPPVRRIDGAYGDRNLVCSCPPVEAFA-------------
Mvan_3131|M.vanbaalenii_PYR-1 FRPKVWPPVRRIDGAYGDRNLVCSCPPVEAFA-------------
MSMEG_3642|M.smegmatis_MC2_155 WRPKVWPPVRRIDGAYGDRNLVCSCPPVEAFA-------------
TH_0290|M.thermoresistible__bu FRPKVWPPVRRIDGAYGDRNLVCSCPPIEAYA-------------
Mb1863|M.bovis_AF2122/97 FRPKVWPAVRRIDGAYGDRNLVCSCPPVEAFA-------------
Rv1832|M.tuberculosis_H37Rv FRPKVWPAVRRIDGAYGDRNLVCSCPPVEAFA-------------
MUL_3050|M.ulcerans_Agy99 FRPKVWPAARRIDGAYGDRNLVCSCPPVEAFA-------------
MAV_2884|M.avium_104 FRPKVWPPVRRIDGAYGDRNLVCSCPPVEAFA-------------
MLBr_02072|M.leprae_Br4923 FRPKVWPPVRRIDGVYGDRNLVCSCLPVEAFV-------------
MAB_2407|M.abscessus_ATCC_1997 FRPKVWPPVRRIDGAYGDRNLVCSCPPIEAFAQ------------
MMAR_0211|M.marinum_M VTDKYWPPVRRIDGVHGDRNLVCSCPSPAAFESTLPPKSASLQVL
* **..*****.:********* . *: