For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTNLWDLKQKVIYSIQWLVVNSLGHQLPRTMLETIRRKTGQPWRTAAGVHPVDNQFGMVCENSEYSDYVY NIKANTAVRTHKSKI
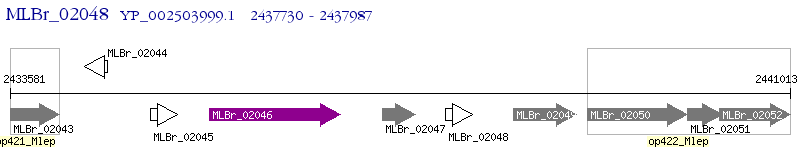
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02048 | - | - | 100% (85) | hypothetical protein MLBr_02048 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1902c | - | 3e-22 | 64.86% (74) | hypothetical protein Mb1902c |
| M. gilvum PYR-GCK | Mflv_4462 | - | 3e-05 | 34.25% (73) | hypothetical protein Mflv_4462 |
| M. tuberculosis H37Rv | Rv1871c | - | 3e-22 | 64.86% (74) | hypothetical protein Rv1871c |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2750 | - | 1e-21 | 63.29% (79) | hypothetical protein MMAR_2750 |
| M. avium 104 | MAV_2845 | - | 4e-20 | 61.64% (73) | hypothetical protein MAV_2845 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3004 | - | 8e-22 | 63.29% (79) | hypothetical protein MUL_3004 |
| M. vanbaalenii PYR-1 | Mvan_1899 | - | 6e-07 | 37.84% (74) | hypothetical protein Mvan_1899 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1902c|M.bovis_AF2122/97 MNAAMNLKREFVHRVQRFVVNPIGRQLP-MTMLETIGRKTGQPRRTAVGG
Rv1871c|M.tuberculosis_H37Rv MNAAMNLKREFVHRVQRFVVNPIGRQLP-MTMLETIGRKTGQPRRTAVGG
MAV_2845|M.avium_104 ----MSLKRRVVHRVQRLLVNPVGRQLP-MTMLETTGRKSGKPRRTAVGG
MMAR_2750|M.marinum_M MAELRDLKRRVVHRMQKLVVNPIGRQLP-VTMLETIGRKSGQPRHTAVGG
MUL_3004|M.ulcerans_Agy99 MAELRDLKRRVVHRMQKLVVNPIGRQLP-VTMLETIGRKSGQPRHTAVGG
MLBr_02048|M.leprae_Br4923 MTNLWDLKQKVIYSIQWLVVNSLGHQLP-RTMLETIRRKTGQPWRTAAGV
Mflv_4462|M.gilvum_PYR-GCK --MAETTRDRVTKFFQKNIANRVMRHIPIQTLLETTGRKSGLPRTTPLGG
Mvan_1899|M.vanbaalenii_PYR-1 -MTADTLRDRVTKFFQKNVANRVMRQMPFQTLLETTGRKSGQPRTTPLGG
: .. .* :.* : :::* *:*** **:* * *. *
Mb1902c|M.bovis_AF2122/97 RVVDNQFWMVSEHGEHSDYVYNIKANPAVRVRIGGRWRSGTAYLLPDDDP
Rv1871c|M.tuberculosis_H37Rv RVVDNQFWMVSEHGEHSDYVYNIKANPAVRVRIGGRWRSGTAYLLPDDDP
MAV_2845|M.avium_104 RVVDNQFWMVSEHGEHSDYVRNIKANPAVRVRVGGRWRTGTAHLLPDDDP
MMAR_2750|M.marinum_M KLVDNQFWMVSEHGEHSDYVRNIKANPAVRVRLDGEWRSGTAHLLPDDDP
MUL_3004|M.ulcerans_Agy99 KLVDNQFWMVSEHGEHSDYVRNIKANPAVRVRLDGEWRSGTAHLLPDDDP
MLBr_02048|M.leprae_Br4923 HPVDNQFGMVCENSEYSDYVYNIKANTAVRT-----HKSKI---------
Mflv_4462|M.gilvum_PYR-GCK RRIGDEFWFVSEFGDRSQYVRNIMADQNVRLRLRGRWHRGIAHLLPEDDA
Mvan_1899|M.vanbaalenii_PYR-1 RRIGDAFWFVSEFGDRSQYIRNIQADPNVRVRLNGRWHRGTAHLLPEDDA
: :.: * :*.* .: *:*: ** *: ** :
Mb1902c|M.bovis_AF2122/97 RQRLRGLPRLNSAGVRAMGTDLLTIRVDLD-
Rv1871c|M.tuberculosis_H37Rv RQRLRGLPRLNSAGVRAMGTDLLTIRVDLD-
MAV_2845|M.avium_104 VQRLGNLPRLNSAMVRLMGSDLLTIRVDLD-
MMAR_2750|M.marinum_M LQRLGNLPRLNSAVVRAIGSELLSIRVDLD-
MUL_3004|M.ulcerans_Agy99 LQRLGNLPRLNSAVVRAIGSELLSIRVDLD-
MLBr_02048|M.leprae_Br4923 -------------------------------
Mflv_4462|M.gilvum_PYR-GCK QARMRMLPRYNNVGVRTFGTNLLTIRVDLID
Mvan_1899|M.vanbaalenii_PYR-1 RARMRTLPGYNNVGVRTFGTNLLTVRVDLID