For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLSLGIVGLPNVGKSTLFNALTRSNVVVANYPFATIEPNEGVVSLPDPRLAKLAELFGSERILPAQVTFV DIAGLVKGASEGAGLGNKFLAHIRECDAICQVVRVFSGDDITHVTGRVDPQSDIGIIDTELILADLDTLE RALPRLEKEARNHKERKPIYAAAINAQQVLNVGTTLFAAGVDGSALRELNLLTNKPFLYVFNADEAVLTA AAWVAELRQLVAPADAVFLDAAIEAELAELDDESAAELLESIGQTERGLDALARAGFRTLALQTFLTVGP KEVRAWTIHQGDTASKAAGVIHTDFEKRFIKAEIVSYDDLIATGSMSAAKAAGKVRTEGKDYVMADGDVV EFRHG*
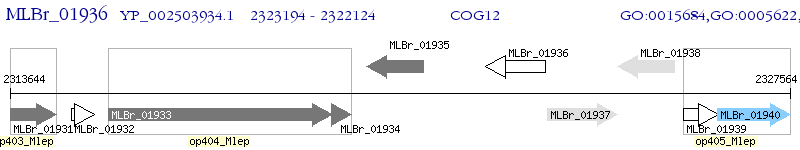
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01936 | - | - | 100% (356) | GTP-dependent nucleic acid-binding protein EngD |
| M. leprae Br4923 | MLBr_01465 | obgE | 2e-15 | 35.16% (128) | GTPase ObgE |
| M. leprae Br4923 | MLBr_00631 | era | 8e-05 | 24.63% (203) | GTP-binding protein Era |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1142 | - | 1e-170 | 85.88% (354) | GTP-dependent nucleic acid-binding protein EngD |
| M. gilvum PYR-GCK | Mflv_2082 | - | 1e-161 | 80.23% (354) | translation-associated GTPase |
| M. tuberculosis H37Rv | Rv1112 | - | 1e-170 | 86.16% (354) | GTP-dependent nucleic acid-binding protein EngD |
| M. abscessus ATCC 19977 | MAB_1266 | - | 1e-157 | 79.10% (354) | GTP-dependent nucleic acid-binding protein EngD |
| M. marinum M | MMAR_4353 | - | 1e-169 | 85.31% (354) | GTP binding protein |
| M. avium 104 | MAV_1232 | ychF | 1e-162 | 83.57% (347) | translation-associated GTPase |
| M. smegmatis MC2 155 | MSMEG_5222 | ychF | 1e-162 | 80.56% (360) | translation-associated GTPase |
| M. thermoresistible (build 8) | TH_1238 | - | 1e-161 | 80.51% (354) | Probable GTP binding protein |
| M. ulcerans Agy99 | MUL_0166 | - | 1e-169 | 85.31% (354) | GTP-dependent nucleic acid-binding protein EngD |
| M. vanbaalenii PYR-1 | Mvan_4625 | - | 1e-162 | 80.79% (354) | translation-associated GTPase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1266|M.abscessus_ATCC_1997 ---MSLNLGIVGLPNVGKSTLFNALTRNNVLAANYPFATIEPNEGVVPLP
MSMEG_5222|M.smegmatis_MC2_155 ---MSLNLGIVGLPNVGKSTLFNALTRNDVLAANYPFATIEPNEGVVPLP
Mflv_2082|M.gilvum_PYR-GCK MDPVGLNLGIVGLPNVGKSTLFNALTRNDVLAANYPFATIEPNEGVVALP
Mvan_4625|M.vanbaalenii_PYR-1 MVPVGLNLGIVGLPNVGKSTLFNALTRNDVLAANYPFATIEPNEGVVALP
TH_1238|M.thermoresistible__bu ---VGLNLGIVGLPNVGKSTLFNALTRNNVLAANYPFATIEPNEGVVPLP
MAV_1232|M.avium_104 ----------MGLPNVGKSTLFNALTRNNVVAANYPFATIEPNEGVVPLP
MMAR_4353|M.marinum_M ---MSLSLGIVGLPNVGKSTLFNALTRNDVLAANYPFATIEPNEGVVSLP
MUL_0166|M.ulcerans_Agy99 ---MSLSLGIVGLPNVGKSTLFNALTRNDVLAANYPFATIEPNEGVVSLP
Mb1142|M.bovis_AF2122/97 ---MSLSLGIVGLPNVGKSTLFNALTRNNVVAANYPFATIEPNEGVVSLP
Rv1112|M.tuberculosis_H37Rv ---MSLSLGIVGLPNVGKSTLFNALTRNNVVAANYPFATIEPNEGVVSLP
MLBr_01936|M.leprae_Br4923 ---MSLSLGIVGLPNVGKSTLFNALTRSNVVVANYPFATIEPNEGVVSLP
:****************.:*:.***************.**
MAB_1266|M.abscessus_ATCC_1997 DPRLDKLAEVFGSARTLPATVTFVDIAGIVKGASEGAGLGNKFLANIREC
MSMEG_5222|M.smegmatis_MC2_155 DPRLDKLAELFGSEKTVPAPVTFVDIAGIVKGASEGAGLGNKFLANIREC
Mflv_2082|M.gilvum_PYR-GCK DPRLTELARMFGSEKIVPAPVTFVDIAGIVKGASEGAGLGNKFLANIRES
Mvan_4625|M.vanbaalenii_PYR-1 DPRLTELARIFGSEKILPAPVTFVDIAGIVKGASEGAGLGNKFLANIREC
TH_1238|M.thermoresistible__bu DPRLEKLAEIFGSEKIVPATVTFVDIAGIVKGASEGAGLGNKFLANIREC
MAV_1232|M.avium_104 DPRLDKLAEMFKSERIVAAPVTFVDIAGLVKGASEGAGLGNKFLAHIREC
MMAR_4353|M.marinum_M DPRLDKLAELFGSERIVAAPVTFVDIAGLVKGASEGAGLGNKFLAHIREC
MUL_0166|M.ulcerans_Agy99 DPRLDKLAELFGSERIVAAPVTFVDIAGLVKGASEGAGLGNKFLAHIREC
Mb1142|M.bovis_AF2122/97 DPRLDKLAELFGSQRVVPAPVTFVDIAGLIKGASEGAGLGNKFLAHIREC
Rv1112|M.tuberculosis_H37Rv DPRLDKLAELFGSQRVVPAPVTFVDIAGLVKGASEGAGLGNKFLAHIREC
MLBr_01936|M.leprae_Br4923 DPRLAKLAELFGSERILPAQVTFVDIAGLVKGASEGAGLGNKFLAHIREC
**** :**.:* * : :.* ********::***************:***.
MAB_1266|M.abscessus_ATCC_1997 DAICQVVRVFADDDVVHVDGRVDPAADIEVIETELILADMQTLEKAVPRL
MSMEG_5222|M.smegmatis_MC2_155 DAICQVVRVFNDDDVVHVDGRVDPRSDIEVIETELILADMQTLEKAVPRL
Mflv_2082|M.gilvum_PYR-GCK DAICQVVRVFADDDVVHVDGRVDPKSDIEVIETELILADMQTLEKAVPRL
Mvan_4625|M.vanbaalenii_PYR-1 DAICQVVRVFADDDVVHVDGRVDPKSDIEVIETELILADMQTLERALPRL
TH_1238|M.thermoresistible__bu DAICQVVRVFSDDDVAHVDGRIDPKADIEVIETELILADLQTLEKALPRL
MAV_1232|M.avium_104 DAICQVVRVFADDDVTHVAGQVDPRSDIEVVETELILADLQTLERAAGRL
MMAR_4353|M.marinum_M DAICQVVRVFHDDDVTHVSGRVDPRSDIEVVETELILADMQTLERAVPRL
MUL_0166|M.ulcerans_Agy99 DAICQVVRVFHDDDVTHVSGRVDPRSDIEVVETELILADMQTLERAVPRL
Mb1142|M.bovis_AF2122/97 DAICQVVRVFVDDDVTHVTGRVDPQSDIEVVETELILADLQTLERATGRL
Rv1112|M.tuberculosis_H37Rv DAICQVVRVFVDDDVTHVTGRVDPQSDIEVVETELILADLQTLERATGRL
MLBr_01936|M.leprae_Br4923 DAICQVVRVFSGDDITHVTGRVDPQSDIGIIDTELILADLDTLERALPRL
********** .**:.** *::** :** :::*******::***:* **
MAB_1266|M.abscessus_ATCC_1997 EKEAKNKKDRQPLLDAAKAAQEVLDGGKTLFS----TGKDWSSVRELNLM
MSMEG_5222|M.smegmatis_MC2_155 EKEARNNKERKPVLEAAVAAQAVLDSGKTLFAGAAAAGIDVTLLRELNLM
Mflv_2082|M.gilvum_PYR-GCK EKEARNNKDRKPVLEAALAAQEILNTGKTLFA----AGMDTTLLRELNLM
Mvan_4625|M.vanbaalenii_PYR-1 EKEARTHKDRRPVYDAAVAAGKILDTGKTLFS----AGFDVSELRELNLL
TH_1238|M.thermoresistible__bu EKEARTHKDRRPTYEAAVAAQEILNSGKTLFA----AGFDATLLRELNLL
MAV_1232|M.avium_104 EKEARTSKERRPVYEAALAAQETLNGGKTLFS----AGTDTSLLRELNLL
MMAR_4353|M.marinum_M EKEARNNKERKPVHEAALAAQEVLDSGKTLFA----AGVDTSLLRELNLL
MUL_0166|M.ulcerans_Agy99 EKEARNNKERKPVHEAALAAQEVLDSGKTLFA----AGVDTSLLRELNLL
Mb1142|M.bovis_AF2122/97 EKEARTNKARKPVYDAALRAQQVLDAGKTLFA----AGVDAAALRELNLL
Rv1112|M.tuberculosis_H37Rv EKEARTNKARKPVYDAALRAQQVLDAGKTLFA----AGVDAAALRELNLL
MLBr_01936|M.leprae_Br4923 EKEARNHKERKPIYAAAINAQQVLNVGTTLFA----AGVDGSALRELNLL
****:. * *:* ** * *: *.***: :* * : :*****:
MAB_1266|M.abscessus_ATCC_1997 TTKPFLYVFNADESVLGDEARVAQLRALVAPADAVFLDAKIESELQELDE
MSMEG_5222|M.smegmatis_MC2_155 TTKPFLYVFNADESVLTDEARQAELREMVAPADAVFLDAKIEAELLELDD
Mflv_2082|M.gilvum_PYR-GCK TTKPFLYVFNADETVLTDEKRVAELRELVAPADAVFLDAKIEAELQELDE
Mvan_4625|M.vanbaalenii_PYR-1 TAKPFLYVFNADEAVLTDTERVAALRELVAPADAVFLDAKIEAELQELDE
TH_1238|M.thermoresistible__bu TTKPFLYVFNCDEEVLTDEARKAELRELVAPADCVFLDAKIEAELQELDD
MAV_1232|M.avium_104 TTKPFLYVFNADESVLTDPARIAELRELVAPADAVFLDAAIESELIELDD
MMAR_4353|M.marinum_M TTKPFLYVFNADESVLTDAARVAQLRELVAPADAVFLDAAIEAELVELDE
MUL_0166|M.ulcerans_Agy99 TTKPFLYVFNADESVLTGAARVAQLRELVAPADAVFLDAAIEAELVELDE
Mb1142|M.bovis_AF2122/97 TTKPFLYVFNADEAVLTDPARVGELRALVAPADAVFLDAAIESELTELDD
Rv1112|M.tuberculosis_H37Rv TTKPFLYVFNADEAVLTDPARVGELRALVAPADAVFLDAAIESELTELDD
MLBr_01936|M.leprae_Br4923 TNKPFLYVFNADEAVLTAAAWVAELRQLVAPADAVFLDAAIEAELAELDD
* ********.** ** . ** :*****.***** **:** ***:
MAB_1266|M.abscessus_ATCC_1997 ESAAELLESIGQAERGLDALARAGFHTLKLQTYLTAGPKEARAWTIHQGD
MSMEG_5222|M.smegmatis_MC2_155 ESAAELLESIGQTERGLDALARAGFHTLKLQTYLTAGPKEARAWTIHQGD
Mflv_2082|M.gilvum_PYR-GCK ESAAELLESIGQTERGLDALARAGFHTLKLQTYLTAGPKEARAWTIHQGD
Mvan_4625|M.vanbaalenii_PYR-1 ESAAELLESIGQTERGLDALARAGFHTLKLQTYLTAGPKEARAWTIHQGD
TH_1238|M.thermoresistible__bu ESARELLESIGQTERGLDALARAGFHTLKLQTFLTAGPKEARAWTIRQGT
MAV_1232|M.avium_104 ESAAELLESIGQTERGLDALARAGFHTLRLQTFLTAGPKEARAWVIHQGD
MMAR_4353|M.marinum_M ESAAELLESIGQTERGLDALARAGFHTLNLQTYLTVGPKEARAWTIHRGD
MUL_0166|M.ulcerans_Agy99 ESAAELLESIGQTERGLDALARAGFHTLNLQTYLTVGPKEARAWTIHRGD
Mb1142|M.bovis_AF2122/97 ESAAELLESIGQSERGLDALARAGFHTLKLQTFLTAGPKEARAWTIHQGD
Rv1112|M.tuberculosis_H37Rv ESAAELLESIGQSERGLDALARAGFHTLKLQTFLTAGPKEARAWTIHQGD
MLBr_01936|M.leprae_Br4923 ESAAELLESIGQTERGLDALARAGFRTLALQTFLTVGPKEVRAWTIHQGD
*** ********:************:** ***:**.****.***.*::*
MAB_1266|M.abscessus_ATCC_1997 TAPKAAGVIHTDFEKGFIKAEVVSYEDLIEAGSMAAAKAAGKVRMEGKEY
MSMEG_5222|M.smegmatis_MC2_155 TAPKAAGVIHSDFEKGFIKAEVVSFDDLVEAGSMAAAKAAGKVRMEGKDY
Mflv_2082|M.gilvum_PYR-GCK TAPKAAGVIHTDFEKGFIKAEVVSFDDLMEAGSMAAAKAAGKVRMEGKDY
Mvan_4625|M.vanbaalenii_PYR-1 TAPKAAGVIHSDFEKGFIKAEVVSFDDLVEAGSMAAAKAAGKVRIEGKDY
TH_1238|M.thermoresistible__bu TAPKAAGVIHSDFEKGFIKAEVVSFDDLVEAGSMAAAKAAGKVRMEGKDY
MAV_1232|M.avium_104 TAPKAAGVIHSDFEKGFIKAEIVSYDDLIAAGSMAAAKAAGKVRMEGKDY
MMAR_4353|M.marinum_M TAPKAAGVIHTDFEKGFIKAEIVSFDDLLAAGSMAAAKAAGKVRMEGKDY
MUL_0166|M.ulcerans_Agy99 TAPKAAGVIHTDFEKGFIKAEIVSFDDLLAAGSMAAAKAAGKVRMEGKDY
Mb1142|M.bovis_AF2122/97 TAPKAAGVIHSDFEKGFIKAEIVSYDDLVAAGSMAAAKAAGKVRIEGKDY
Rv1112|M.tuberculosis_H37Rv TAPKAAGVIHSDFEKGFIKAEIVSYDDLVAAGSMAAAKAAGKVRIEGKDY
MLBr_01936|M.leprae_Br4923 TASKAAGVIHTDFEKRFIKAEIVSYDDLIATGSMSAAKAAGKVRTEGKDY
**.*******:**** *****:**::**: :***:********* ***:*
MAB_1266|M.abscessus_ATCC_1997 VMVDGDVVEFRFNV---------
MSMEG_5222|M.smegmatis_MC2_155 VMADGDVVEFRHGGTSGSGKK--
Mflv_2082|M.gilvum_PYR-GCK VMADGDVVEFRFNV---------
Mvan_4625|M.vanbaalenii_PYR-1 VMADGDVVEFRFNV---------
TH_1238|M.thermoresistible__bu VMQDGDVVEFRFNV---------
MAV_1232|M.avium_104 VMADGDVVEFRFNV---------
MMAR_4353|M.marinum_M VMADGDVVEFRFQTSSGGKAAGR
MUL_0166|M.ulcerans_Agy99 VMADGDVVEFRFRTSSGGKAAGR
Mb1142|M.bovis_AF2122/97 VMADGDVVEFRFNV---------
Rv1112|M.tuberculosis_H37Rv VMADGDVVEFRFNV---------
MLBr_01936|M.leprae_Br4923 VMADGDVVEFRHG----------
** ********.