For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIARLMTTSPELVTGKAFPNVVVTGIAMTTALATDAETTWKLLLDNQSGIRMLDDPFIEEFNLPVRIGGH LLEEFDHQLTRVELRRMGYLQRMSTVLSRRLWENAGSPEVDTNRLMVSIGTGLGSAEELVFSYDDMRARG MKAVSPLAVQKYMPNGAAAAVGLEHHAKAGVMTPVSACASGSEAIAHAWQQIVLGEADSAICGGVETKIE AVPIAGFSQMRIVMSTKNDNPAGACRPFDRDRDGFVFGEAGALMLIETEDSAKARSANILARIMGASITS DGFHMVAPDPNGERAGHAIARAVHLAGLSPSDIDHVNAHATGTQVGDLAEAKAINKALCNNRPAVYAPKS ALGHSVGAVGAVESILTVLALRDQVIPPTLNLVNLDPDIDLDVVAGKPRPGDYRYAVNNSFGFGGHNVAI AFGCY
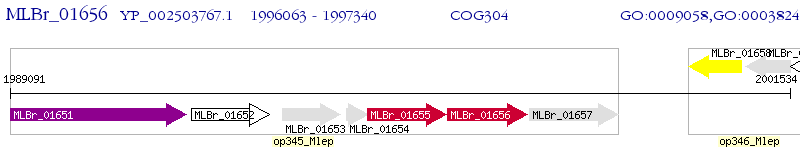
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01656 | kasB | - | 100% (425) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. leprae Br4923 | MLBr_01655 | kasA | e-156 | 66.27% (418) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. leprae Br4923 | MLBr_00101 | pks13 | 1e-37 | 28.74% (435) | polyketide synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2270 | kasB | 0.0 | 90.78% (423) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. gilvum PYR-GCK | Mflv_2776 | - | 0.0 | 78.16% (412) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. tuberculosis H37Rv | Rv2246 | kasB | 0.0 | 90.78% (423) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. abscessus ATCC 19977 | MAB_1877c | - | 1e-157 | 66.35% (419) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. marinum M | MMAR_3339 | kasB | 0.0 | 88.67% (415) | 3-oxoacyl- |
| M. avium 104 | MAV_2191 | - | 0.0 | 85.25% (427) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. smegmatis MC2 155 | MSMEG_4328 | - | 0.0 | 78.26% (414) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. thermoresistible (build 8) | TH_2837 | - | 0.0 | 81.23% (405) | PUTATIVE beta-ketoacyl synthase |
| M. ulcerans Agy99 | MUL_1303 | kasB | 0.0 | 88.67% (415) | 3-oxoacyl-(acyl carrier protein) synthase II |
| M. vanbaalenii PYR-1 | Mvan_3757 | - | 0.0 | 78.64% (412) | 3-oxoacyl-(acyl carrier protein) synthase II |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2776|M.gilvum_PYR-GCK ---------------------MASPTTGN-GLPDVVVTGIAMTTALATDA
Mvan_3757|M.vanbaalenii_PYR-1 ---------------------MASPTTGN-GLPDVVVTGIAMTTSLATDA
Mb2270|M.bovis_AF2122/97 MGVPPLAGASRTDMEGTFARPMTELVTGK-AFPYVVVTGIAMTTALATDA
Rv2246|M.tuberculosis_H37Rv MGVPPLAGASRTDMEGTFARPMTELVTGK-AFPYVVVTGIAMTTALATDA
MLBr_01656|M.leprae_Br4923 -------------MIARLMTTSPELVTGK-AFPNVVVTGIAMTTALATDA
MMAR_3339|M.marinum_M ---------------------MTELVTGK-AFPHVVVTGIAMTTALATDA
MUL_1303|M.ulcerans_Agy99 ---------------------MTELVTGK-AFPHVVVTGIAMTTALATDA
MAV_2191|M.avium_104 MRAATARKVAGQVREGIFARPMTELVTGK-TLPNVVVTGIAMTTALATDA
MSMEG_4328|M.smegmatis_MC2_155 ---------------------MAALSTGN-GLPNVVVTGVAMTTALATDA
TH_2837|M.thermoresistible__bu ----------------------------------VVVTGVAMTTALATDA
MAB_1877c|M.abscessus_ATCC_199 ---------------------MTLPSTANGGFPNVVVTALAATTSIAPDI
****.:* **::*.*
Mflv_2776|M.gilvum_PYR-GCK EQTWTSLLDGRSGIRKLDDPFVEQFDLPVRIGGHLLEEFDAQLTKVENRR
Mvan_3757|M.vanbaalenii_PYR-1 EATWKSLLDGHSGIRKLQDPFVEEFDLPVQIGGHLLEEFDSQLTKVENRR
Mb2270|M.bovis_AF2122/97 ETTWKLLLDRQSGIRTLDDPFVEEFDLPVRIGGHLLEEFDHQLTRIELRR
Rv2246|M.tuberculosis_H37Rv ETTWKLLLDRQSGIRTLDDPFVEEFDLPVRIGGHLLEEFDHQLTRIELRR
MLBr_01656|M.leprae_Br4923 ETTWKLLLDNQSGIRMLDDPFIEEFNLPVRIGGHLLEEFDHQLTRVELRR
MMAR_3339|M.marinum_M ETTWKLLLDSQSGIRTLDDPFVEEFNLPVRIGGHLLEEFDHQLTRVELRR
MUL_1303|M.ulcerans_Agy99 ETTWKLLLDSQSGIRTLDDPFVEEFNLPVRIGGHLLEEFDHQLTRVELRR
MAV_2191|M.avium_104 ETTWKLLLDSQSGIRTLDDPFVEEFDLPVRIGGHLLEEFDSQLTRVELRR
MSMEG_4328|M.smegmatis_MC2_155 ESTWKKLLDGQSGIRTLTDPFVEEYNLPVRIGGHLLEEFDSELTRVELRR
TH_2837|M.thermoresistible__bu EGTWKKLLDGQSGIRLLEDDFIEKYDLPVRIGGHLLEDFDSELTRVELRR
MAB_1877c|M.abscessus_ATCC_199 EGTWKGLIAGESGIEVLEDEFVEKFGLPVWIGGHLKEPIDNHMTRLELRR
* **. *: .***. * * *:*::.*** ***** * :* .:*::* **
Mflv_2776|M.gilvum_PYR-GCK MSYIQKMATVLGRRVWENAGAPE-VDTERLLVSIGTGLGGAEALVFAYDG
Mvan_3757|M.vanbaalenii_PYR-1 MSYVQKMATVLGRRVWENAGSPE-VDTERLLVSIGTGLGGAEALVFAYDG
Mb2270|M.bovis_AF2122/97 MGYLQRMSTVLSRRLWENAGSPE-VDTNRLMVSIGTGLGSAEELVFSYDD
Rv2246|M.tuberculosis_H37Rv MGYLQRMSTVLSRRLWENAGSPE-VDTNRLMVSIGTGLGSAEELVFSYDD
MLBr_01656|M.leprae_Br4923 MGYLQRMSTVLSRRLWENAGSPE-VDTNRLMVSIGTGLGSAEELVFSYDD
MMAR_3339|M.marinum_M TAYLQRMSTVIGRRVWENAGSPE-VDTNRLAVSIGTGLGSAEELVFGYDE
MUL_1303|M.ulcerans_Agy99 TAYLQRMSTVIGRRVWENAGSPE-VDTNRLAVSIGTGLGSAEELVFGYDE
MAV_2191|M.avium_104 TGYLQRMSTILGRRVWENAGSPE-VDSNRLMVSIGTGLGSSEEMVFSYDD
MSMEG_4328|M.smegmatis_MC2_155 LSYLQKMATVLGRRVWANAGSPE-VDARRLMVSMGTGLGSTEELVFAYDG
TH_2837|M.thermoresistible__bu LSYLQKMSTVLSRRVWENAGSPE-VDPRRLMVSIGTGMGSTEELVFSYDD
MAB_1877c|M.abscessus_ATCC_199 MAYVQRMALHVGRQVWQNAGSPDDIDKDRLGVVIGTGLGGGERIVETYDT
.*:*:*: :.*::* ***:*: :* ** * :***:*. * :* **
Mflv_2776|M.gilvum_PYR-GCK LRERGLRSVSPLSVQMYMPNGPSAVVGLERHAKAGVVTPVSACASGSEGI
Mvan_3757|M.vanbaalenii_PYR-1 LRERGLRSVSPLSVQMYMPNGPSAVVGLERHAKAGVVTPVSACASGSEGI
Mb2270|M.bovis_AF2122/97 MRARGMKAVSPLTVQKYMPNGAAAAVGLERHAKAGVMTPVSACASGAEAI
Rv2246|M.tuberculosis_H37Rv MRARGMKAVSPLTVQKYMPNGAAAAVGLERHAKAGVMTPVSACASGAEAI
MLBr_01656|M.leprae_Br4923 MRARGMKAVSPLAVQKYMPNGAAAAVGLEHHAKAGVMTPVSACASGSEAI
MMAR_3339|M.marinum_M MRVRGFRAISPLAVQKYMPNGAAAAVGLERHAKAGVMTPVSACASGSEAI
MUL_1303|M.ulcerans_Agy99 MRVRGFRAISPLAVQKYMPNGAAAAVGLERHAKAGVMTPVSACASGSEAI
MAV_2191|M.avium_104 MRARGMKAVSPLGVQKYMPNGAAAAVGLEHHAKAGVITPVSACASGSEGI
MSMEG_4328|M.smegmatis_MC2_155 MRSKGLRAVSPLVVQMYMPNGAAAAIGLERQAKAGVCTSISACASGSEAI
TH_2837|M.thermoresistible__bu MRAKGLKAVSPLAVQKYMPNAAAAAVGLERKARAGVITPVSACASGSEGI
MAB_1877c|M.abscessus_ATCC_199 MNEHGIRKVSPLAVQMCMPNGAAAVVGLEIGARAGVITPVSACSSGSEAI
:. :*:: :*** ** ***..:*.:*** *:*** *.:***:**:*.*
Mflv_2776|M.gilvum_PYR-GCK AQAWRNIVMGEADIAICGGVEQRIEAVPIAAFAQMRIVMSTNNDDPAGAC
Mvan_3757|M.vanbaalenii_PYR-1 AQAWRNIVLGEADIAICGGVEQRIEAVPIAGFAQMRIVMSANNDDPAGAC
Mb2270|M.bovis_AF2122/97 ARAWQQIVLGEADAAICGGVETRIEAVPIAGFAQMRIVMSTNNDDPAGAC
Rv2246|M.tuberculosis_H37Rv ARAWQQIVLGEADAAICGGVETRIEAVPIAGFAQMRIVMSTNNDDPAGAC
MLBr_01656|M.leprae_Br4923 AHAWQQIVLGEADSAICGGVETKIEAVPIAGFSQMRIVMSTKNDNPAGAC
MMAR_3339|M.marinum_M ARAWQQVVLGEADIAICGGVETRIEAVPIAGFANMRIVMSTNNDDPAGAC
MUL_1303|M.ulcerans_Agy99 ARAWQQVVLGEADIAICGGVETRIEAVPIAGFANMRIVMSTNNDDPAGAC
MAV_2191|M.avium_104 AQAWRNIVFGEADIAICGGVETKIEAVPIAAFAQMRIVMSTKNDDPVGAC
MSMEG_4328|M.smegmatis_MC2_155 ANAWRNIVLGEADIAICGGVETKIEAVPIAGFAQMRIVLSNSNDDPEGAC
TH_2837|M.thermoresistible__bu AHAWRNIVLGEADIAICGGVETKIEAVPIAGFAQMRIVLSTTNDDPPGAC
MAB_1877c|M.abscessus_ATCC_199 AHAWRQIVMGDADMIVCGGVEGMIEALPIAAFSMMR-AMSTRNDEPEKAS
*.**:::*:*:** :***** ***:***.*: ** .:* **:* *.
Mflv_2776|M.gilvum_PYR-GCK RPFDKDRDGFVFGEAGALLVIETEEHAKARGATPLARLMGASITSDGFHM
Mvan_3757|M.vanbaalenii_PYR-1 RPFDKDRDGFVFGEAGALMVIETEEHARARGATPLARVMGASITSDGYHM
Mb2270|M.bovis_AF2122/97 RPFDRDRDGFVFGEGGALLLIETEEHAKARGANILARIMGASITSDGFHM
Rv2246|M.tuberculosis_H37Rv RPFDRDRDGFVFGEGGALLLIETEEHAKARGANILARIMGASITSDGFHM
MLBr_01656|M.leprae_Br4923 RPFDRDRDGFVFGEAGALMLIETEDSAKARSANILARIMGASITSDGFHM
MMAR_3339|M.marinum_M RPFDKDRDGFVFGEGGALMVIETEEHAKARGANILARIMGASITSDGFHM
MUL_1303|M.ulcerans_Agy99 RPFDKDRDGFVFGEGGALMVIETEEHAKARGANILARIMGASITSDGFHM
MAV_2191|M.avium_104 RPFDRDRTGFVFGEAGALMVIETEEHAKARGANILARIMGASITSDGYHM
MSMEG_4328|M.smegmatis_MC2_155 KPFDKDRNGFVFGEAGALMVIETEEHAKARGANILGRLMGASVTSDGYHI
TH_2837|M.thermoresistible__bu KPFDKDRNGFVFGEAGALMVIETEEHAKARGANILGRLMGASITSDGYHI
MAB_1877c|M.abscessus_ATCC_199 RPFDKDRDGFVFGEAGALMVIETEEHAKARGAQIIGRVMGAGITSDAYHM
:***:** ******.***::****: *:**.* :.*:***.:***.:*:
Mflv_2776|M.gilvum_PYR-GCK VAPDPDGERAGYAMTRAIELAGLTPTDIDHVNAHATGTSVGDVAEGRAIN
Mvan_3757|M.vanbaalenii_PYR-1 VAPDPNGERAGYAITRAIELAGLKPTDIDHVNAHATGTSVGDLAEGHAIN
Mb2270|M.bovis_AF2122/97 VAPDPNGERAGHAITRAIQLAGLAPGDIDHVNAHATGTQVGDLAEGRAIN
Rv2246|M.tuberculosis_H37Rv VAPDPNGERAGHAITRAIQLAGLAPGDIDHVNAHATGTQVGDLAEGRAIN
MLBr_01656|M.leprae_Br4923 VAPDPNGERAGHAIARAVHLAGLSPSDIDHVNAHATGTQVGDLAEAKAIN
MMAR_3339|M.marinum_M VAPDPNGERAGHAITRAVQLAGLQPSDIDHVNAHATGTQVGDVAEGRAIN
MUL_1303|M.ulcerans_Agy99 VAPDPNGERAGHAITRAVQLAGLQPSDIDHVNAHATGTQVGDVAEGRAIN
MAV_2191|M.avium_104 VAPDPNGERAGHAMSRAIQLAGLTPGDIDHVNAHATGTSVGDVAESKAIN
MSMEG_4328|M.smegmatis_MC2_155 VAPDPNGEQAGYAMTRAIELAGLKPTDIDHVNAHATGTNVGDVAESKAIN
TH_2837|M.thermoresistible__bu VAPDPNGEQAGWAMTRAIELAGLQPSDIDHVNAHATGTTVGDVAEGKAIN
MAB_1877c|M.abscessus_ATCC_199 VAPGPDGIRAGAAMTRAIQTAGLSPKDIDHINAHGTATPIGDVAEANAIH
***.*:* :** *::**:. *** * ****:***.*.* :**:**..**:
Mflv_2776|M.gilvum_PYR-GCK RALGGH--RPAVYAPKSALGHSVGAVGAVESILTVQALRDGVVPPTLNLK
Mvan_3757|M.vanbaalenii_PYR-1 LAMGGH--RPAVYAPKSALGHSVGAVGAVESILTVQSLRDGIIPPTLNLK
Mb2270|M.bovis_AF2122/97 NALGGN--RPAVYAPKSALGHSVGAVGAVESILTVLALRDQVIPPTLNLV
Rv2246|M.tuberculosis_H37Rv NALGGN--RPAVYAPKSALGHSVGAVGAVESILTVLALRDQVIPPTLNLV
MLBr_01656|M.leprae_Br4923 KALCNN--RPAVYAPKSALGHSVGAVGAVESILTVLALRDQVIPPTLNLV
MMAR_3339|M.marinum_M NALGSH--KPAVYAPKSALGHSVGAVGAVESILTVLALRDQVVPPTLNLV
MUL_1303|M.ulcerans_Agy99 NALGSH--KPAVYAPKSALGHSVGAVGAVESILTVLALRDQVVPPTLNLV
MAV_2191|M.avium_104 NALGPHGGNAAVYAPKAALGHSVGAVGAVESILTVLALRDQVVPPTLNLE
MSMEG_4328|M.smegmatis_MC2_155 NAMGGH--KPAVYAPKGALGHSVGAVGAVESILTVLALRDGIIPPTRNLN
TH_2837|M.thermoresistible__bu NAMKGH--RPAVYAPKAALGHSVGAVGAVESILTVFALRDGVIPPTLNLK
MAB_1877c|M.abscessus_ATCC_199 VAGVQH---AAVYAPKSALGHSIGAVGALESVLTVLALRDGVIPPTLNLD
* : .******.*****:*****:**:*** :*** ::*** **
Mflv_2776|M.gilvum_PYR-GCK NQDPEIDLDVVAGSPRPGNYNYALNNSFGFGGHNVALAFGKY
Mvan_3757|M.vanbaalenii_PYR-1 NLDPEIDLDVVAGGPRPGNYEYAINNSFGFGGHNVALAFGKY
Mb2270|M.bovis_AF2122/97 NLDPEIDLDVVAGEPRPGNYRYAINNSFGFGGHNVAIAFGRY
Rv2246|M.tuberculosis_H37Rv NLDPEIDLDVVAGEPRPGNYRYAINNSFGFGGHNVAIAFGRY
MLBr_01656|M.leprae_Br4923 NLDPDIDLDVVAGKPRPGDYRYAVNNSFGFGGHNVAIAFGCY
MMAR_3339|M.marinum_M NLDPEIDLDVVAGKPRPGNYRYAVNNSFGFGGHNVAITFGRY
MUL_1303|M.ulcerans_Agy99 NLDPEIDLDVVAGKPRPGNYRYAVNNSFGFGGHNVAITFGRY
MAV_2191|M.avium_104 NLDPEIDLDVVAGKPRPGNYQYAINNSFGFGGHNVAIAFGRY
MSMEG_4328|M.smegmatis_MC2_155 NLDPEIDLDVVAGEPRQGDYKYAINNSFGFGGHNVALAFGKY
TH_2837|M.thermoresistible__bu NQDPEIDLDVVAGEPRTGDFKYAINNSFGFGGHNVALAFGKY
MAB_1877c|M.abscessus_ATCC_199 NLDPEIDLDVVAGEPRHGDFKYAINNSFGFGGHNVALAFGKY
* **:******** ** *::.**:************::** *