For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RHAKSSYPRGFPDHIADHDRRLAPRGVREASLAGGWLRTNVPAIEKVLCSTAMRARETLTHSGIEAPVRY TERLYRADPDTVIKEIKAISDEVTTSLIVSHEPTISAVALALTGSGTNNDAAQRISTKFPTSGIAVLNVA GRWQHLELESAELVAFHVPR*
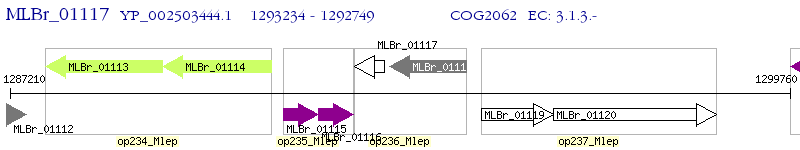
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01117 | - | - | 100% (161) | hypothetical protein MLBr_01117 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1307c | - | 2e-54 | 64.81% (162) | hypothetical protein Mb1307c |
| M. gilvum PYR-GCK | Mflv_2254 | - | 3e-37 | 45.78% (166) | putative phosphohistidine phosphatase, SixA |
| M. tuberculosis H37Rv | Rv1276c | - | 6e-55 | 65.43% (162) | hypothetical protein Rv1276c |
| M. abscessus ATCC 19977 | MAB_1419c | - | 1e-38 | 50.92% (163) | putative phosphoglycerate/bisphosphoglycerate mutase |
| M. marinum M | MMAR_4144 | sixA | 2e-52 | 62.96% (162) | phosphohistidine phosphatase SixA |
| M. avium 104 | MAV_1425 | - | 6e-48 | 60.49% (162) | phosphoglycerate mutase family protein |
| M. smegmatis MC2 155 | MSMEG_5006 | - | 5e-43 | 54.55% (165) | phosphohistidine phosphatase |
| M. thermoresistible (build 8) | TH_1851 | - | 2e-41 | 53.94% (165) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4007 | sixA | 3e-53 | 63.58% (162) | phosphohistidine phosphatase SixA |
| M. vanbaalenii PYR-1 | Mvan_4437 | - | 1e-42 | 53.01% (166) | putative phosphohistidine phosphatase, SixA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2254|M.gilvum_PYR-GCK MVSVRHSYCRAALRQVCGRAPACPTVNGMSDRLRTLLLLRHAKSDYP---
Mvan_4437|M.vanbaalenii_PYR-1 ----------------------------MSDRHRTLVLLRHAKSDYP---
MSMEG_5006|M.smegmatis_MC2_155 --------------------------------MATLLLMRHAKSDYP---
TH_1851|M.thermoresistible__bu ----------------------------MSDPFRTLVLLRHGKSAYP---
Mb1307c|M.bovis_AF2122/97 --------------------------------------MRHAKSAYP---
Rv1276c|M.tuberculosis_H37Rv --------------------------------------MRHAKSAYP---
MLBr_01117|M.leprae_Br4923 --------------------------------------MRHAKSSYPRGF
MMAR_4144|M.marinum_M -----------------------------MSDRHTLLLMRHAKSQYP---
MUL_4007|M.ulcerans_Agy99 -----------------------------MSDRHTLLLMRHAKSQYP---
MAV_1425|M.avium_104 -----------------------------MTGRRTLLLMRHAKSDYP---
MAB_1419c|M.abscessus_ATCC_199 ---------------------------MPDDAKRTLILMRHAKSAYP---
:**.** **
Mflv_2254|M.gilvum_PYR-GCK -DGVVDHDRPLASRGVREAALAGDWIRSTIGDIDAVLCSTATRTRQTLEQ
Mvan_4437|M.vanbaalenii_PYR-1 -DGVVDHDRPLAPRGVREGALAGEWIRANVGAVDAVLCSTATRTRQTLER
MSMEG_5006|M.smegmatis_MC2_155 -DGVVDHERPLAPRGVREAALAGDWIRANAPGIDAVLCSTATRTRQTLAR
TH_1851|M.thermoresistible__bu -PDCADRDRPLAERGIREAGLAGDWLRAHVPPVDAVLCSPATRTRQTFDR
Mb1307c|M.bovis_AF2122/97 -DGIADHDRPLAPRGIREAGLAGGWLRANLPAVDAVLCSTATRARQTLAH
Rv1276c|M.tuberculosis_H37Rv -DGIADHDRPLAPRGIREAGLAGGWLRANLPAVDAVLCSTATRARQTLAH
MLBr_01117|M.leprae_Br4923 PDHIADHDRRLAPRGVREASLAGGWLRTNVPAIEKVLCSTAMRARETLTH
MMAR_4144|M.marinum_M -PGVADHERPLAPRGRREAGLAGDWLRGNAPAVDEVLCSTAIRTRETLAR
MUL_4007|M.ulcerans_Agy99 -PGVADHERPLAPRGRREAGLAGDWLRGNAPAVDEVLCSTAIRTRETLAR
MAV_1425|M.avium_104 -DGVADHDRPLAPRGIRQAGLAGDWLRAGAPAIDAVLCSTATRTRETLRN
MAB_1419c|M.abscessus_ATCC_199 -PGVDDHQRPLNLRGTRQAAQAGTWLQDHFNRIDAVLCSTATRTRATLTQ
*::* * ** *:.. ** *:: :: ****.* *:* *: .
Mflv_2254|M.gilvum_PYR-GCK TGITAP-VQFVDRIYDATAGILIEEVNGAQARFGEDVSTLLVIGHEPVMS
Mvan_4437|M.vanbaalenii_PYR-1 TGITAP-VQYVDRIYDATPGIVIEEINDVQSRFGEVVSTLLVIGHEPVMS
MSMEG_5006|M.smegmatis_MC2_155 TGIDAP-TEYVEKIYDAVPGTVIGEINDVVSRFGTQPDTVLVVGHEPAMS
TH_1851|M.thermoresistible__bu TRLTAP-VHYAERLYDATPGTVIDVINGVAAHFDTDIRTLLVIGHEPALS
Mb1307c|M.bovis_AF2122/97 TGIDAP-ARYAERLYGAAPGTVIEEIN----RVGDNVTSLLVVGHEPTTS
Rv1276c|M.tuberculosis_H37Rv TGIDAP-ARYAERLYGAAPGTVIEEIN----RVGDNVTTLLVVGHEPTTS
MLBr_01117|M.leprae_Br4923 SGIEAP-VRYTERLYRADPDTVIKEIK----AISDEVTTSLIVSHEPTIS
MMAR_4144|M.marinum_M SGIDAP-VHYSQRLYGAVPGTLIGEIN----RIGEDVATLLVVGHEPTMS
MUL_4007|M.ulcerans_Agy99 SGIDAP-VHYSQRLYGAVPGTLIGEIN----RIGEDVATLLVVGHEPTMS
MAV_1425|M.avium_104 THIEAP-VRYSERLYASTPGIVIDEIN----TVGDDVSTLLVIGHEPTMS
MAB_1419c|M.abscessus_ATCC_199 TGIHSQLVRFSDKLYDSTPGITLGEIN----AVPDSAVTVLVVGHEPTTS
: : : ..: :::* : .. : :: . : *::.***. *
Mflv_2254|M.gilvum_PYR-GCK SVALGLADERSGNGPVADKISAKYPTSSVAVLRSSAPWEQWALRGAELID
Mvan_4437|M.vanbaalenii_PYR-1 ALALSLADEPSSDTPAYQQISAKFPTSAVAVLRTATPWASLALRGAELVD
MSMEG_5006|M.smegmatis_MC2_155 SVALGLDDG--ANVAVAEEISQKFPTSAIAVLRFDGPWDELALGGAQLVT
TH_1851|M.thermoresistible__bu RVALGLSDG--GNATAAQRISEKFPTSAIAVLRTAADWDELALQGAALVA
Mb1307c|M.bovis_AF2122/97 ALAIVLASISGTDAAVAERISEKFPTSGIAVLRVAGHWADVEPGCAALVG
Rv1276c|M.tuberculosis_H37Rv ALAIVLASISGTDAAVAERISEKFPTSGIAVLRVAGHWADVEPGCAALVG
MLBr_01117|M.leprae_Br4923 AVALALTG-SGTNNDAAQRISTKFPTSGIAVLNVAGRWQHLELESAELVA
MMAR_4144|M.marinum_M EVALGLAGATGTDAAAAERLSAKFPTSAIAVLQVAGRWEDLELGGAALIT
MUL_4007|M.ulcerans_Agy99 AVALGLAGATGTDAAAAERLSAKFPTSAIAVLQVAGRWEDLELGGAALIT
MAV_1425|M.avium_104 ALALGLGGGRGANPAAAERISNKFPTSAIAVLAVPCRWTELELGAATLSD
MAB_1419c|M.abscessus_ATCC_199 QLALALADAGTSSAEAANRIATKFPTSAIAVLTTRSPWARLELGGASLTT
:*: * . . . :.:: *:***.:*** * * *
Mflv_2254|M.gilvum_PYR-GCK FHTAR
Mvan_4437|M.vanbaalenii_PYR-1 FHVPR
MSMEG_5006|M.smegmatis_MC2_155 FHVPR
TH_1851|M.thermoresistible__bu FHVPR
Mb1307c|M.bovis_AF2122/97 FHVPR
Rv1276c|M.tuberculosis_H37Rv FHVPR
MLBr_01117|M.leprae_Br4923 FHVPR
MMAR_4144|M.marinum_M FHVPR
MUL_4007|M.ulcerans_Agy99 FHVPR
MAV_1425|M.avium_104 FVVAR
MAB_1419c|M.abscessus_ATCC_199 FHVPR
* ..*