For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SFPDATITRLPTVLQPYAQRYHELIKFAIVGGTTFIIDSAIFYTLKLTILEPKPVTAKVVAGIVAVIASY VLNREWSFRDRGGRERHNEALLFFAFSGIGVLLSMAPLWFSSYVLQLRAPTVSLTVENLADFLSAYIIGN LLQMAFRFWAFRRWVFPDAFARNPEKTLESALTAGGIAEVFEDAIDGVFEDFGDALLRAWRNRSRRLDLS PASQLGDSSEPRVSKTS*
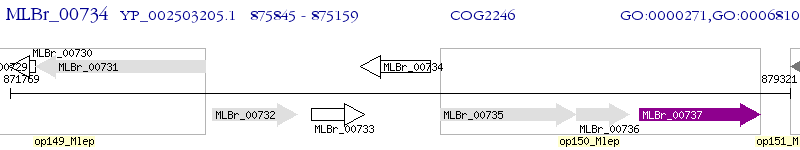
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00734 | - | - | 100% (228) | hypothetical protein MLBr_00734 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3305 | - | 1e-108 | 84.58% (227) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_4748 | - | 2e-87 | 70.82% (233) | GtrA family protein |
| M. tuberculosis H37Rv | Rv3277 | - | 1e-109 | 84.65% (228) | transmembrane protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1259 | - | 1e-100 | 84.40% (218) | hypothetical protein MMAR_1259 |
| M. avium 104 | MAV_4247 | - | 1e-104 | 83.48% (230) | hypothetical protein MAV_4247 |
| M. smegmatis MC2 155 | MSMEG_1817 | - | 5e-91 | 73.68% (228) | transmembrane protein |
| M. thermoresistible (build 8) | TH_0888 | - | 3e-92 | 73.25% (228) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_2629 | - | 1e-105 | 83.98% (231) | hypothetical protein MUL_2629 |
| M. vanbaalenii PYR-1 | Mvan_1712 | - | 2e-88 | 72.81% (228) | GtrA family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1259|M.marinum_M --------------------------------------------------
MUL_2629|M.ulcerans_Agy99 -------------------------------------------------M
MLBr_00734|M.leprae_Br4923 -------------------------------------------------M
Mb3305|M.bovis_AF2122/97 MNEVTAGVRELATAIMVSRHLTGVLAGHGSQTVTYHFASILCSSVHSLVV
Rv3277|M.tuberculosis_H37Rv MNEVTAGVRELATAIMVSRHLTGVLAGHGSQTVTYHFASILCSSVHSLVV
MAV_4247|M.avium_104 -------------------------------------------------M
Mflv_4748|M.gilvum_PYR-GCK -------------------------------------------------M
Mvan_1712|M.vanbaalenii_PYR-1 --------------------------------MTARCGIWAPGSVNWLLV
MSMEG_1817|M.smegmatis_MC2_155 -------------------------------------------------M
TH_0888|M.thermoresistible__bu -------------------------------------------------V
MMAR_1259|M.marinum_M ------------MVQPFAQRHHELIKFAIVGGTTFIIDSAIFYTLKLTIL
MUL_2629|M.ulcerans_Agy99 SFADATIARLPRVVQPFAQRHHELIKFAIVGGTTFIIDSAIFYTLKLTIL
MLBr_00734|M.leprae_Br4923 SFPDATITRLPTVLQPYAQRYHELIKFAIVGGTTFIIDSAIFYTLKLTIL
Mb3305|M.bovis_AF2122/97 SFADATIARLPGVVQPYAQRHHELIKFAIVGGTTFIIDTAIFYTLKLTVL
Rv3277|M.tuberculosis_H37Rv SFADATIARLPGVVQPYAQRHHELIKFAIVGGTTFIIDTAIFYTLKLTVL
MAV_4247|M.avium_104 SFAEATIARLPRLLQPYLLRHHELIKFAIVGGTTFVIDSAIFYTLKLTIL
Mflv_4748|M.gilvum_PYR-GCK SFADATIRRLPGPIRPYAERHHELIKFAIVGATTFVIDSAIFFTLKLTIL
Mvan_1712|M.vanbaalenii_PYR-1 SFADATIRRLPGPIRPYAERHHELIKFAIVGATTFVIDSAIFFTLKLTIL
MSMEG_1817|M.smegmatis_MC2_155 SFADATIARLPRFIRPFAERHHELIKFAIVGATTFVIDSAIFYTLKLTVL
TH_0888|M.thermoresistible__bu SFADATIARLPRPIRPYAERHHELIKFAIVGATTFIIDSAIFYTLKLTVL
::*: *:**********.***:**:***:*****:*
MMAR_1259|M.marinum_M EPKPVTAKVIAGIVAVIASYVLNREWSFRDRGGRERHHEALLFFAFSGVG
MUL_2629|M.ulcerans_Agy99 EPKPVTAKVIAGIVAVIASYVLNREWSFRDRGGRERHHEALLFFAFSGVG
MLBr_00734|M.leprae_Br4923 EPKPVTAKVVAGIVAVIASYVLNREWSFRDRGGRERHNEALLFFAFSGIG
Mb3305|M.bovis_AF2122/97 EPKPVTAKVIAGIVAVIASYVLNREWSFRDRGGRERHHEALLFFAFSGVG
Rv3277|M.tuberculosis_H37Rv EPKPVTAKVIAGIVAVIASYVLNREWSFRDRGGRERHHEALLFFAFSGVG
MAV_4247|M.avium_104 EPKPVTAKVIAGIVAVIASYVLNREWSFRNRGGRERHHEALLFFAFSGVG
Mflv_4748|M.gilvum_PYR-GCK EPKPVTAKIIAGIVAVIASYILNREWSFRDRGGRERHHEALLFFGVSGVG
Mvan_1712|M.vanbaalenii_PYR-1 EPKPVTAKIIAGIVAVIASYILNREWSFRDRGGRERHHEALLFFGVSGVG
MSMEG_1817|M.smegmatis_MC2_155 EPKPVTAKIIAGIVAVIASYILNREWSFRDRGGRERHHEAFLFFAVSGVG
TH_0888|M.thermoresistible__bu EPKPVTAKIIAGIVAVIASYILNREWSFRDRGGRERHHEALLFFGVSGVG
********::**********:********:*******:**:***..**:*
MMAR_1259|M.marinum_M VLLSMAPLWFSSYVLQLRVPMVSLTVENLADFVSAYIIGNLLQMAFRFWA
MUL_2629|M.ulcerans_Agy99 VLLSMAPLWFSSYVLQLRVPMVSLTVENLADFVSAYIIGNLLQMAFRFWA
MLBr_00734|M.leprae_Br4923 VLLSMAPLWFSSYVLQLRAPTVSLTVENLADFLSAYIIGNLLQMAFRFWA
Mb3305|M.bovis_AF2122/97 VLLSMAPLWFSSYILQLRVPTVSLTMENIADFISAYIIGNLLQMAFRFWA
Rv3277|M.tuberculosis_H37Rv VLLSMAPLWFSSYILQLRVPTVSLTMENIADFISAYIIGNLLQMAFRFWA
MAV_4247|M.avium_104 VLLSMAPLWFSSYVLQLRQPTVSLTVENVADFISAYIIGNLLQMAFRFWA
Mflv_4748|M.gilvum_PYR-GCK VLLSMAPLWVSSYVLMLRVPMVSLMTENIADFVSAYIIGNLLQMAFRFWA
Mvan_1712|M.vanbaalenii_PYR-1 VLLSMAPLWVSSYVLMLRVPMVSLAAENVADFISAYIIGNLLQMAFRFWA
MSMEG_1817|M.smegmatis_MC2_155 VLLSMAPLWISSYVLMLRVPEVSLTTENIADFISAYIIGNLLQMAFRFWA
TH_0888|M.thermoresistible__bu VVLSMAPLWISSYVLMLRAPMVSLTVENVADFISAYIIGNLLQMAFRFWA
*:*******.***:* ** * *** **:***:*****************
MMAR_1259|M.marinum_M FRRWVFPDEFARNPDKALESALTAGGIAEVFEDEIEGG----NVT--LLR
MUL_2629|M.ulcerans_Agy99 FRRWVFPDEFARNPDKALESALTAGGIAEVFEDEIEGG----NVT--LLR
MLBr_00734|M.leprae_Br4923 FRRWVFPDAFARNPEKTLESALTAGGIAEVFEDAIDGVFEDFGDA--LLR
Mb3305|M.bovis_AF2122/97 FRRWVFPDEFARNPDKALESALTAGGIAEVFEDVLEGGFEDGNVT--LLR
Rv3277|M.tuberculosis_H37Rv FRRWVFPDEFARNPDKALESALTAGGIAEVFEDVLEGGFEDGNVT--LLR
MAV_4247|M.avium_104 FRRWVFPDQFARDPDKALESALTAGGIAEIFEDAFEDD--GGNVT--LLR
Mflv_4748|M.gilvum_PYR-GCK FRRFVFPDEFARNPEKALESTLTGGGLAEALEDEYELRHPHGPEPKGNVT
Mvan_1712|M.vanbaalenii_PYR-1 FRRFVFPDEFARNPEKALESTLTGGGFAEVLEDEYELRHPSGPD---NVT
MSMEG_1817|M.smegmatis_MC2_155 FRRFVFPDEFARNPDKALESTLTGGGLAEALEDEYEVSH--GPDS--VVT
TH_0888|M.thermoresistible__bu FRRWVFPDEFGRAPDKALESTFTAGGLAEALEDHLEAHSERSSN----VT
***:**** *.* *:*:***::*.**:** :** : :
MMAR_1259|M.marinum_M SWRNKSARANRSDARSAAQLGASSEPRVSKTS
MUL_2629|M.ulcerans_Agy99 SWRNKSARANRSDARSAAQLGASSEPRVSKTS
MLBr_00734|M.leprae_Br4923 AWRNRSRRLDLS---PASQLGDSSEPRVSKTS
Mb3305|M.bovis_AF2122/97 AWRNRANR--------FAQLGDSSEPRVSKTL
Rv3277|M.tuberculosis_H37Rv AWRNRANR--------FAQLGDSSEPRVSKTS
MAV_4247|M.avium_104 AWRNRAGR--------LTQLGDSSEPRVSKTS
Mflv_4748|M.gilvum_PYR-GCK PMR-MSRRR-----RNQRQRNDSDEPRVSKTS
Mvan_1712|M.vanbaalenii_PYR-1 PMR-PSRRR-----WKQRP-SDPSEPRVSKTS
MSMEG_1817|M.smegmatis_MC2_155 PMR-RSRGR-----GAPPQLGDSSDPRVSKTS
TH_0888|M.thermoresistible__bu PLHGRARLRSG---RHSRQLGDSSDPRVSKTS
. : : . ..:******