For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LNLMIFKVGDTVVYPHHGAALVEAIETRTINGEQKEYLVLKVAQGDLTVRVPAENAEYVGVRDVVGQEGL DQVFQVLRAPHTEEPTNWSRRYKANLEKLASGDVNKVSEVVRDLWRRDQDRGLSAGEKRMLAKARQILVG ELALAESTDDAKAETILDEVLAAAS
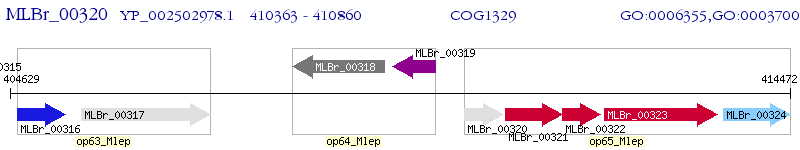
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00320 | - | - | 100% (165) | putative transcription factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3614c | - | 4e-86 | 97.53% (162) | putative transcription factor |
| M. gilvum PYR-GCK | Mflv_1444 | - | 1e-84 | 95.06% (162) | CarD family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3583c | - | 4e-86 | 97.53% (162) | transcription factor |
| M. abscessus ATCC 19977 | MAB_0568 | - | 1e-80 | 91.36% (162) | putative CarD-like transcriptional regulator |
| M. marinum M | MMAR_5083 | - | 6e-86 | 97.53% (162) | transcriptional regulatory protein |
| M. avium 104 | MAV_0570 | - | 5e-86 | 97.53% (162) | transcriptional regulator, CarD family protein |
| M. smegmatis MC2 155 | MSMEG_6077 | - | 4e-85 | 95.68% (162) | transcriptional regulator, CarD family protein |
| M. thermoresistible (build 8) | TH_1183 | - | 7e-75 | 95.86% (145) | POSSIBLE TRANSCRIPTION FACTOR |
| M. ulcerans Agy99 | MUL_4159 | - | 4e-86 | 97.53% (162) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5340 | - | 3e-84 | 94.44% (162) | CarD family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1444|M.gilvum_PYR-GCK ---MIFKVGDTVVYPHHGAALIEAIETRTIKGEQKEYLVLKVAQGDLTVR
Mvan_5340|M.vanbaalenii_PYR-1 ---MIFKVGDTVVYPHHGAALIEAIETRTIKGEQKEYLVLKVSQGDLTVR
MSMEG_6077|M.smegmatis_MC2_155 ---MIFKVGDTVVYPHHGAALIEAIETRTIKGEQKEYLVLKVAQGDLTVR
MAV_0570|M.avium_104 ---MIFKVGDTVVYPHHGAALVEAIETRTIKGEQKEYLVLKVAQGDLTVR
MUL_4159|M.ulcerans_Agy99 ---MIFKVGDTVVYPHHGAALVEAIETRTIKGEQKEYLVLKVAQGDLTVR
MMAR_5083|M.marinum_M ---MIFKVGDTVVYPHHGAALVEAIETRTIKGEQKEYLVLKVAQGDLTVR
Rv3583c|M.tuberculosis_H37Rv ---MIFKVGDTVVYPHHGAALVEAIETRTIKGEQKEYLVLKVAQGDLTVR
Mb3614c|M.bovis_AF2122/97 ---MIFKVGDTVVYPHHGAALVEAIETRTIKGEQKEYLVLKVAQGDLTVR
MLBr_00320|M.leprae_Br4923 MNLMIFKVGDTVVYPHHGAALVEAIETRTINGEQKEYLVLKVAQGDLTVR
TH_1183|M.thermoresistible__bu --------------------LIEAIETRTIKGEQKEYLVLKVAQGDLTVR
MAB_0568|M.abscessus_ATCC_1997 ---MIFKVGDTVVYPHHGAALIEAIETRTIKGEQKEYLVLKVAQGDLTVR
*:********:***********:*******
Mflv_1444|M.gilvum_PYR-GCK VPADNAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
Mvan_5340|M.vanbaalenii_PYR-1 VPADNAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MSMEG_6077|M.smegmatis_MC2_155 VPADNAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MAV_0570|M.avium_104 VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MUL_4159|M.ulcerans_Agy99 VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MMAR_5083|M.marinum_M VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
Rv3583c|M.tuberculosis_H37Rv VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
Mb3614c|M.bovis_AF2122/97 VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MLBr_00320|M.leprae_Br4923 VPAENAEYVGVRDVVGQEGLDQVFQVLRAPHTEEPTNWSRRYKANLEKLA
TH_1183|M.thermoresistible__bu VPAENAEYVGVRDVVGQEGLDKVFQVLRAPHTEEPTNWSRRYKANLEKLA
MAB_0568|M.abscessus_ATCC_1997 VPAENAEYVGVRDVVGREGLDTVFQVLRAPHTEEPTNWSRRFKANQEKLA
***:************:**** *******************:*** ****
Mflv_1444|M.gilvum_PYR-GCK SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAENTDD
Mvan_5340|M.vanbaalenii_PYR-1 SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAENTDD
MSMEG_6077|M.smegmatis_MC2_155 SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAENTDD
MAV_0570|M.avium_104 SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAESTDD
MUL_4159|M.ulcerans_Agy99 SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAESTDD
MMAR_5083|M.marinum_M SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAESTDD
Rv3583c|M.tuberculosis_H37Rv SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAESTDD
Mb3614c|M.bovis_AF2122/97 SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAESTDD
MLBr_00320|M.leprae_Br4923 SGDVNKVSEVVRDLWRRDQDRGLSAGEKRMLAKARQILVGELALAESTDD
TH_1183|M.thermoresistible__bu SGDVNKVAEVVRDLWRRDQERGLSAGEKRMLAKARQILVGELALAENTDD
MAB_0568|M.abscessus_ATCC_1997 SGDVKKVAEVVRDLWRREQERGLSAGEKRMLAKARQILVGELALAENTNA
****:**:*********:*:**************************.*:
Mflv_1444|M.gilvum_PYR-GCK AKASTILDEVLAAAS
Mvan_5340|M.vanbaalenii_PYR-1 AKASTILDEVLAAAS
MSMEG_6077|M.smegmatis_MC2_155 AKAETILDEVLAAAS
MAV_0570|M.avium_104 AKAETILDEVLAAAS
MUL_4159|M.ulcerans_Agy99 AKAETILDEVLAAAS
MMAR_5083|M.marinum_M AKAETILDEVLAAAS
Rv3583c|M.tuberculosis_H37Rv AKAETILDEVLAAAS
Mb3614c|M.bovis_AF2122/97 AKAETILDEVLAAAS
MLBr_00320|M.leprae_Br4923 AKAETILDEVLAAAS
TH_1183|M.thermoresistible__bu AKAETILDEVLAAAS
MAB_0568|M.abscessus_ATCC_1997 AKADTILDEVLAAAS
***.***********