For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVLTVLVFALTESFTTSWIKQFSIAGILWVVLIGQLTGELFVRDFAAAERPDDVVKTQLFARIVTLLTWI
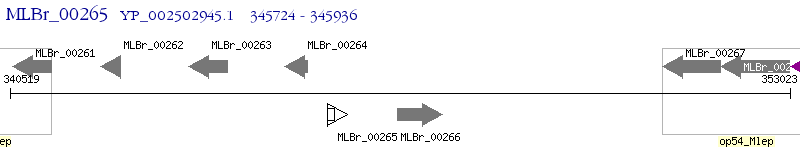
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00265 | - | - | 100% (70) | hypothetical protein MLBr_00265 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3941 | - | 3e-14 | 49.28% (69) | hypothetical protein Mflv_3941 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4417 | - | 4e-15 | 53.62% (69) | hypothetical protein MMAR_4417 |
| M. avium 104 | MAV_1182 | - | 1e-14 | 54.29% (70) | hypothetical protein MAV_1182 |
| M. smegmatis MC2 155 | MSMEG_3475 | - | 6e-13 | 44.29% (70) | hypothetical protein MSMEG_3475 |
| M. thermoresistible (build 8) | TH_0418 | - | 5e-11 | 38.57% (70) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2458 | - | 2e-13 | 44.29% (70) | hypothetical protein Mvan_2458 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3475|M.smegmatis_MC2_155 ---MGILFGFAPWIIYWVLVGNVPFLTAVLIALATAVATFVISRISGTPG
TH_0418|M.thermoresistible__bu LPTVSILFGFAPWIVFWVLVGNVPFTAAVLVALAIAVATLVIGHRTGAPG
Mflv_3941|M.gilvum_PYR-GCK ---MGIIFGLAPWLVYWVLVGNVPFHTAVLVALAVAVASFVVARVSGSPG
Mvan_2458|M.vanbaalenii_PYR-1 ---MGIIFGFAPWIVYWVLVGNVPFHAAVLIALAVAVAAFVAARVARSPG
MMAR_4417|M.marinum_M -----MLFGFSPWLVYWVLVGNVPFETAVAVALLIALAVLAVGRSAQKPG
MAV_1182|M.avium_104 ---MGMLFGLAPWIVYWVLVGNVPFVAAVLVALALAAVSLGLGGVLRRRW
MLBr_00265|M.leprae_Br4923 --------------------------------------------------
MSMEG_3475|M.smegmatis_MC2_155 RTLEIGALATFLVLTVLTFVLSQDAMERWMQPLSNAGIFLVALVGAIIGK
TH_0418|M.thermoresistible__bu RALQIGAVSTFVVLTVVTVVLHQTFLERWLQPLGNAGILLAALTGVLVDK
Mflv_3941|M.gilvum_PYR-GCK RVLEIGAVATFAVLTVLTFTVSGSFLEQWLQPLSNAGILVAALVGVLAGR
Mvan_2458|M.vanbaalenii_PYR-1 RTLEIGAVATFLVLAALTFTMSSAFMERWLQPLSNAGIFLVALVGALVGR
MMAR_4417|M.marinum_M RTVEIGSVATFSVLTVLTFALSEAFLARWIVAVSNFGIFLVAVLGVLMGK
MAV_1182|M.avium_104 QLFDFASVAVLLVLAVLAFTLSESILQRWILPLSNAGIFLAALAGMLIGK
MLBr_00265|M.leprae_Br4923 -----------MVLTVLVFALTESFTTSWIKQFSIAGILWVVLIGQLTGE
**:.:...: *: .. **: ..: * : ..
MSMEG_3475|M.smegmatis_MC2_155 PFVKEYAEVGQPPGVVESDLFKRIVAILTWVWVGAFAGMTVSSAIPPIVQ
TH_0418|M.thermoresistible__bu PFVREYAAVGQPPEVLESDVFNKITTVLTWVWIGAFAGMTVSSAIPPIVQ
Mflv_3941|M.gilvum_PYR-GCK PFVREFAVVGQPKEVVESEPFARITSVLTWIWIAAFAGMTVSSAIPPMVQ
Mvan_2458|M.vanbaalenii_PYR-1 PFVREFAVVGQPKEVIESEPFARITSTLTWIWIAAFGGMTVSSAIPPLVQ
MMAR_4417|M.marinum_M PLVREYAAAELPSDVVNTELFKRMTMILTWIWVAAFAGMTLSSLVPPIVE
MAV_1182|M.avium_104 PFVAEFAAAEQAPDVAKTELFGRIVKILGWIWVATFAGMTVSSAIPSVLQ
MLBr_00265|M.leprae_Br4923 LFVRDFAAAERPDDVVKTQLFARIVTLLTWI-------------------
:* ::* . . * ::: * ::. * *:
MSMEG_3475|M.smegmatis_MC2_155 G-----DATILDTKTPLSFVCYWVIPFALLGVAALVSRVLPDRMMEGIND
TH_0418|M.thermoresistible__bu G-----DATLLDVNTPLSFVFYWVIPFSLLGLAALASRLLPDRLAEEIGN
Mflv_3941|M.gilvum_PYR-GCK G-----DATILDTQTPLSFIFYWVVPFALLGVAALLTRILPDRMVPPAEE
Mvan_2458|M.vanbaalenii_PYR-1 G-----DATILDTRTPLSFLCYWVVPFSLLGVAALLTRILPDRMLPPEEE
MMAR_4417|M.marinum_M S-----DATILDTKTPVSFMCYWVIPVSLLGLAALASQLLPTRMLAGIDD
MAV_1182|M.avium_104 GSAGAAPAYMLDTKTPLSFLCYWIIPFALLGLAAVVSRLLPDRMLIGIDD
MLBr_00265|M.leprae_Br4923 --------------------------------------------------
MSMEG_3475|M.smegmatis_MC2_155 VVRKTTFVAYSEAAIDELYYLAQEHANREVGPGQEAYDVRVGGSGTPLVG
TH_0418|M.thermoresistible__bu APRTTSFVAYSEATIDELYYLAQEHANREVGAGKEAYDVRVGGKGTPLVG
Mflv_3941|M.gilvum_PYR-GCK IVRKTTFVAFAEAEIDQLIYLATEHANREAGAGKEAYDVRIGSKGVPLVG
Mvan_2458|M.vanbaalenii_PYR-1 IVRKTTFVAFAEAEIDQLIYLATEHANREVGAGKEAYDVRIGSKGVPLVG
MMAR_4417|M.marinum_M VVRETSFVAYDEATIDELYYLAQEHANREVGPGKEAYNVKVGGMGTPLTG
MAV_1182|M.avium_104 LERETSFVAYDEATIDELYFLAQEHANREVGPGKEAYAVKVGGMGTPLTG
MLBr_00265|M.leprae_Br4923 --------------------------------------------------
MSMEG_3475|M.smegmatis_MC2_155 DESRMSWPSTYKVRDRRR
TH_0418|M.thermoresistible__bu DESRMSWQSTYKVRDRRR
Mflv_3941|M.gilvum_PYR-GCK DETRESWPSTYKVRDRKR
Mvan_2458|M.vanbaalenii_PYR-1 DDTRESWPSTYKVRERKR
MMAR_4417|M.marinum_M DEARKSWPSTYKVRDKRR
MAV_1182|M.avium_104 DESRKSWPSTYKVRDKRR
MLBr_00265|M.leprae_Br4923 ------------------