For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIERLGNLGDAPRVLRSLGDATTGLDLPPPAALTGEWFDALAIIAPSLSVQPVDVNDAFAVQLDARTPDS AVAVGGGWVGYLSYPDPAADKQPNRIPEAAGGWTDCVLRRDRGAQWWYESLSGAPMPGWLAAALATTPAP ARNCQVDWERTDWGAHRDGVLACLEAIRAGEIYQACVCTQFTGTVTGAPLDFFIAVVARTAPARAAYIAG PWGAVASLSPELFLRRRGAGVTSSPIKGTLPLAAWPSALQASAKEVAENIMIVDLVRNDLGRVAITGTVT VPELLVVRHAPGVWHLVSTVTAQVPVELPMAALLDATFPPASITGTPKLRARKLISQWEHRRRGIYCGTV GLASPIAGCELNVAIRTVEFDTAGNAVLGVGGGITADSDPDAELQECLHKAAPILRLASVATVASVG*
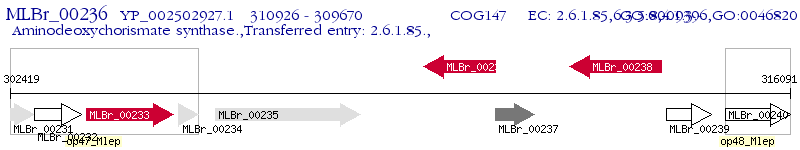
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00236 | pabB | - | 100% (418) | aminodeoxychorismate synthase component I |
| M. leprae Br4923 | MLBr_01269 | trpE | 7e-29 | 31.31% (313) | anthranilate synthase component I |
| M. leprae Br4923 | MLBr_00808 | entC | 2e-14 | 31.55% (206) | putative isochorismate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1032c | pabB | 0.0 | 79.13% (412) | aminodeoxychorismate synthase component I |
| M. gilvum PYR-GCK | Mflv_1926 | - | 1e-158 | 71.18% (406) | para-aminobenzoate synthase component I |
| M. tuberculosis H37Rv | Rv1005c | pabB | 0.0 | 79.13% (412) | aminodeoxychorismate synthase component I |
| M. abscessus ATCC 19977 | MAB_1124c | - | 1e-138 | 60.92% (412) | aminodeoxychorismate synthase component I |
| M. marinum M | MMAR_4488 | pabD | 0.0 | 77.12% (424) | para-aminobenzoate synthase component, PabD |
| M. avium 104 | MAV_1127 | pabB | 0.0 | 77.27% (418) | para-aminobenzoate synthase component I |
| M. smegmatis MC2 155 | MSMEG_5446 | pabB | 1e-151 | 66.01% (409) | para-aminobenzoate synthase component I |
| M. thermoresistible (build 8) | TH_1145 | pabB | 1e-154 | 67.96% (412) | para-aminobenzoate synthase, component I |
| M. ulcerans Agy99 | MUL_4660 | pabD | 0.0 | 76.42% (424) | aminodeoxychorismate synthase component I |
| M. vanbaalenii PYR-1 | Mvan_4807 | - | 1e-156 | 67.96% (412) | para-aminobenzoate synthase component I |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1926|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4807|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4488|M.marinum_M -------------------------------------MACENPSFCQAKA
MUL_4660|M.ulcerans_Agy99 -------------------------------------MACENPSFCQAKA
MLBr_00236|M.leprae_Br4923 --------------------------------------------------
Mb1032c|M.bovis_AF2122/97 MALAARTATPPGTETGAGHGSNLAWELSTRTKSPRSHLRCENPQFCQART
Rv1005c|M.tuberculosis_H37Rv --------------------MNLAWELSTRTKSPRSHLRCENPQFCQART
MAV_1127|M.avium_104 --------------------------------------------------
MSMEG_5446|M.smegmatis_MC2_155 --------------------------------------------------
TH_1145|M.thermoresistible__bu --------------------------------------------------
MAB_1124c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1926|M.gilvum_PYR-GCK MRIERLGDLG---DAPSVLRALGVATAHLGLAPPAALIGDWFGSRAVIAP
Mvan_4807|M.vanbaalenii_PYR-1 MRIERLGDLG---SAPTVLRTLAAAASASGIAPPAALLGDWFSSRAVIAP
MMAR_4488|M.marinum_M VLIDRLGDLG---SAPQVLRAVGHATRQLDLPPPAALTGEWFGALAVIAP
MUL_4660|M.ulcerans_Agy99 VLIDRLGDLG---SAPQVLRAVGHATRQLDLPPPAALTGEWFGALAVIAP
MLBr_00236|M.leprae_Br4923 MRIERLGNLG---DAPRVLRSLGDATTGLDLPPPAALTGEWFDALAIIAP
Mb1032c|M.bovis_AF2122/97 VRIDRLGDLG---GAPAVLRAVGRATSRLDLPPPAALTGEWFGALAVIAP
Rv1005c|M.tuberculosis_H37Rv VRIDRLGDLG---GAPAVLRAVGRATSRLDLPPPAALTGEWFGALAVIAP
MAV_1127|M.avium_104 MRIDRLGDLG---PAPQVLRAVGDVTRRLGLPPPAALTGDWFDALAVIAP
MSMEG_5446|M.smegmatis_MC2_155 MRIERFCTPDGQADAPSVLRSVAAAASAAGLAPPAALIGDWFGSRAVIAP
TH_1145|M.thermoresistible__bu VRIEALGELG---PATAVLRTLEAATAARGLPPPAALIGDWFGARAVIAP
MAB_1124c|M.abscessus_ATCC_199 MHVQQLGDLG---DPPAVLRSLAAAARDTQRPAPAALIGSWYGSRAIIAP
: :: : . .. ***:: .: ..**** *.*:.: *:***
Mflv_1926|M.gilvum_PYR-GCK SVSVSAVAPPTVFDVTPGYRCDG-----AVGGGWFGYLSYPD-AGADGVG
Mvan_4807|M.vanbaalenii_PYR-1 SLIAGEVAPRDAFAVTPGSRCDG-----PVGGGWFGYLSYPD-GGADGLG
MMAR_4488|M.marinum_M SVSVRPVTSTDTFPIRSGADTAAASVPGAVGGGWVGYLSYPD-PGADGHG
MUL_4660|M.ulcerans_Agy99 SVSVRPVTSTDTFPIRRGADTAAASVPGAVGGGWVGYLSYPD-PGADGHG
MLBr_00236|M.leprae_Br4923 SLSVQPVDVNDAFAVQLDARTPDSAV--AVGGGWVGYLSYPD-PAADKQP
Mb1032c|M.bovis_AF2122/97 SVSIQPVSGDDVFSGPPGTGGPDATG--AVGGGWVGYLSYPD-AGADGRP
Rv1005c|M.tuberculosis_H37Rv SVSIQPVSGDDVFSGPPGTGGPDATG--AVGGGWVGYLSYPD-AGADGRP
MAV_1127|M.avium_104 SLPVRPVPPGDVFAVG-TARTPQPPH---VGGGWIGYLSYPD-GGADAKP
MSMEG_5446|M.smegmatis_MC2_155 TVTALPAEPDAVFDVPQGSAEGAA-----VGGGWFGYLSYPD-PGACESA
TH_1145|M.thermoresistible__bu SVAVEPVAPDAVFAGPTG-VGGTA-----VGGGWFGYLSXPDSPDAGDRS
MAB_1124c|M.abscessus_ATCC_199 SLATRPVLPRQAFGGKASHVGTVAPG--AVGGGWIGYLGYPD-RAATGRA
:: . .* *****.***. ** *
Mflv_1926|M.gilvum_PYR-GCK P-----RIPEAAGGWSDSVLRCDRDGAWWHESLSGATMSGWIEQALRSPA
Mvan_4807|M.vanbaalenii_PYR-1 P-----RIPQAAGGWADTVLRCDRDGVWWHESLSGAALPGWVTEAVQSVP
MMAR_4488|M.marinum_M G-----RIPEAAGGWTDSVLRRDRDGQWWYESLSGAPMPDWLRNALSATP
MUL_4660|M.ulcerans_Agy99 G-----RIPEAAGGWTDSVLRRERDGQWWYENLSGAPMPDWLGNALSATP
MLBr_00236|M.leprae_Br4923 N-----RIPEAAGGWTDCVLRRDRGAQWWYESLSGAPMPGWLAAALATTP
Mb1032c|M.bovis_AF2122/97 H-----RIPEAAGGWTDCVLRRDRDGQWWYESLSGAPIADWLASALATTR
Rv1005c|M.tuberculosis_H37Rv H-----RIPEAAGGWTDCVLRRDRDGQWWYESLSGAPIADWLASALATTR
MAV_1127|M.avium_104 N-----RIPEAAGGWTDCVLRRDRDGQWWYESLTGAPMPRWLAGALAAPA
MSMEG_5446|M.smegmatis_MC2_155 P-----RIPVAAGGWSDCVLRQDTEGQWWYESLAGAPLPAWLRDLAPTP-
TH_1145|M.thermoresistible__bu PESRAARLPEAAGGWSDCVLRQDRDGRWWYESLTGTTVPDWVVAALRTPQ
MAB_1124c|M.abscessus_ATCC_199 Q-----RLPAAAGGWTNAVLRLDEDECWWYESLLDQECPEWILHALTHPC
*:* *****:: *** : **:*.* . . *:
Mflv_1926|M.gilvum_PYR-GCK E-----PGRATVTWEPADRDAHRLGVLACLDAIAAGEVYQACVCTQFTGV
Mvan_4807|M.vanbaalenii_PYR-1 D-----PRDAVVTWGEPDRHAHRRGVLACLEAIAAGEVYQACVCTQFAGV
MMAR_4488|M.marinum_M GLSTRPAATCLIDWGTADRHAHREGVLACLDAIAAGEVYQACVCTQFTGT
MUL_4660|M.ulcerans_Agy99 GLSARPAATCLIDWGTADRHAHREGVLACLDAIAAGEVYQACVCTQFTGT
MLBr_00236|M.leprae_Br4923 ----APARNCQVDWERTDWGAHRDGVLACLEAIRAGEIYQACVCTQFTGT
Mb1032c|M.bovis_AF2122/97 ASVARPAPACRIDWEPADRAAHRDGVLACLEAIGAGEVYQACVCTQFAGT
Rv1005c|M.tuberculosis_H37Rv ASVARPAPACRIDWEPADRAAHRDGVLACLEAIGAGEVYQACVCTQFAGT
MAV_1127|M.avium_104 G----PPRPCRIDWDVADRAAHRDGVLACLEAIRAGEVYQACVCTQFRGT
MSMEG_5446|M.smegmatis_MC2_155 ------SRPHTVDWVPPDRGAHRRGVLACLEAIAAGEVYQACVCTRFTGR
TH_1145|M.thermoresistible__bu P-----SRPHIVRWTPPDRXAHRFGVTACLDAIGAGEIYQACVCTQFVGR
MAB_1124c|M.abscessus_ATCC_199 E-----PAPWSIDWTAPDREHHRGAVGRCLEAIAAGEVYQACVCTQFRGS
. : * .* ** .* **:** ***:*******:* *
Mflv_1926|M.gilvum_PYR-GCK LDGDPLEFFTDAVTRTAPARAAYVAGDWGAVASLSPELFLRRCGDAVASS
Mvan_4807|M.vanbaalenii_PYR-1 LAGAPVDFFAEAAARTVPARAAFVAGDWGAVASLSPELFLRRCGEAVTSS
MMAR_4488|M.marinum_M VAGDPLDFFIDGVARTSPARAAYVAGDWGAIASLSPELFLQRRGDTVASS
MUL_4660|M.ulcerans_Agy99 VAGDPLDFFIDGVARTSPARAAYVAGDWGAIASLSPELFLQRRGDTVTSS
MLBr_00236|M.leprae_Br4923 VTGAPLDFFIAVVARTAPARAAYIAGPWGAVASLSPELFLRRRGAGVTSS
Mb1032c|M.bovis_AF2122/97 VTGSPLDFFIDGFGRTAPSRSAFVAGPWGAVASLSPELFLRRRGSVVTSS
Rv1005c|M.tuberculosis_H37Rv VTGSPLDFFIDGFGRTAPSRSAFVAGPWGAVASLSPELFLRRRGSVVTSS
MAV_1127|M.avium_104 VGGAPLDFFVDGVARTDPARAAYVEGEWGAVASLSPELFLRRRGTVVTSS
MSMEG_5446|M.smegmatis_MC2_155 LDGSPSDFFADAVARTAPARAAYVAGDWGAVASLSPELFLSRRGSSVSSS
TH_1145|M.thermoresistible__bu IDGAPIDFFADTVARTGPARAAYLAGRWGAVASLSPELFLRRTGEHIASS
MAB_1124c|M.abscessus_ATCC_199 LTGKALDFFADAVTRTAPARAAYLAGIWGAVASLSPELFLRRNGTDVCSS
: * . :** ** *:*:*:: * ***:********* * * : **
Mflv_1926|M.gilvum_PYR-GCK PIKGTLPRSADPAALRASVKDVAENIMIVDLVRNDLGRVARTGSVAVPEL
Mvan_4807|M.vanbaalenii_PYR-1 PIKGTLPDFADPATLRASVKDVAENIMIVDLVRNDLGRVARTGSVSVPEL
MMAR_4488|M.marinum_M PIKGTLPLDAWPSALRASPKEVAENIMIVDLVRNDLGRVAITGTVTVPEL
MUL_4660|M.ulcerans_Agy99 PIKGTLPLDAWPSALRASPKEVAENIMIVDLVRNDLGRVAITGTVTVPEL
MLBr_00236|M.leprae_Br4923 PIKGTLPLAAWPSALQASAKEVAENIMIVDLVRNDLGRVAITGTVTVPEL
Mb1032c|M.bovis_AF2122/97 PIKGTLPLDAPPSALRASAKEVAENIMIVDLVRNDLGRVAVTGTVTVPEL
Rv1005c|M.tuberculosis_H37Rv PIKGTLPLDAPPSALRASAKEVAENIMIVDLVRNDLGRVAVTGTVTVPEL
MAV_1127|M.avium_104 PIKGTLPLDAWPSALRASAKEVAENIMIVDLVRNDLGRVATVGTVSVPEL
MSMEG_5446|M.smegmatis_MC2_155 PIKGTLPRHADPAGLRASVKDVAENVMIVDLVRNDLGRVARTGSVTVPEL
TH_1145|M.thermoresistible__bu PIKGTLPRDADPAALRASVKDVAENIMIVDLVRNDLGRIARTGSVTVPEL
MAB_1124c|M.abscessus_ATCC_199 PIKGTLAATEDPAALLTSIKDVAENVMITDLVRNDLGRIADTGTVTVPEL
******. *: * :* *:****:**.*********:* .*:*:****
Mflv_1926|M.gilvum_PYR-GCK LAVRPAPGVWHLVSTVTAQVPSELPTSALLDATFPPASVTGTPKLRAREL
Mvan_4807|M.vanbaalenii_PYR-1 LAVRPAPGVWHLVSTVSARVPEDTPTAVLLDATFPPASVTGTPKSRARGL
MMAR_4488|M.marinum_M LVVRRAPGVWHLVSTVSAQVPVELPTSALLDATFPPASVTGTPKLRARQL
MUL_4660|M.ulcerans_Agy99 LVVRRAPGVWHLVSTVSAQVPVELPTSALLDATFPPASVTGTPKLRARQL
MLBr_00236|M.leprae_Br4923 LVVRHAPGVWHLVSTVTAQVPVELPMAALLDATFPPASITGTPKLRARKL
Mb1032c|M.bovis_AF2122/97 LVVRPAPGVWHLVSTVSARVPLEEPMSALLDAAFPPASVTGTPKLRARQL
Rv1005c|M.tuberculosis_H37Rv LVVRPAPGVWHLVSTVSARVPLEEPMSALLDAAFPPASVTGTPKLRARQL
MAV_1127|M.avium_104 LVVRRAPGVWHLVSTVSAQVPTELPTSALLDAAFPPASVTGTPKHRARQL
MSMEG_5446|M.smegmatis_MC2_155 LAVKPAPGVWHLVSTVAAEVEQCVPMGALLDAAFPPASVTGTPKGRAREL
TH_1145|M.thermoresistible__bu LAVRPAPGVWHLVSTVAARVDPAVPMAAVLAAAFPPASVTGTPKLRARQL
MAB_1124c|M.abscessus_ATCC_199 LAVHPAPGVWHLVSTVAAKVPDEATAGDLLAAAFPPASVTGTPKTRARQL
*.*: ***********:*.* . . :* *:*****:***** *** *
Mflv_1926|M.gilvum_PYR-GCK LTGWERNRRGIYCGTVGLASPSAGCELNVAIRTVEFAAERTAVLGVGGGI
Mvan_4807|M.vanbaalenii_PYR-1 LTGWEPHRRGIYCGTVGLASPVAGCELNVAIRTVEFGADGSAVLGVGGGI
MMAR_4488|M.marinum_M ISQWEQRRRGIYCGTVGLASPIAGCELNVAIRTVEFDTVGNAVLGVGGGI
MUL_4660|M.ulcerans_Agy99 ISQWEQRRRGIYCGTVGLVSPIAGCELNVAIRTVEFDTIGNAVLGVGGGI
MLBr_00236|M.leprae_Br4923 ISQWEHRRRGIYCGTVGLASPIAGCELNVAIRTVEFDTAGNAVLGVGGGI
Mb1032c|M.bovis_AF2122/97 ISQWERYRRGIYCGTVGLASPVAGCELNVAIRTVEFDTAGNAVLGVGGGI
Rv1005c|M.tuberculosis_H37Rv ISQWERYRRGIYCGTVGLASPVAGCELNVAIRTVEFDTAGNAVLGVGGGI
MAV_1127|M.avium_104 LSKWEPATRGIYCGTIGLASPVAGCELNVAIRTVEFDAAGGAVLGVGGGI
MSMEG_5446|M.smegmatis_MC2_155 LRAWEPGLRGIYCGTVGLASPVAGCELNVAIRTVEFGADGSAVLGVGGGI
TH_1145|M.thermoresistible__bu LSRWEPFRRGVYCGTVGLASPVAGCELNVAIRTVEFDPHGAAVLGVGGGI
MAB_1124c|M.abscessus_ATCC_199 LRQWEAEHRGVYCGTIGMLSPAAGLELNVAIRTVEIAPDGSAVLGVGGGI
: ** **:****:*: ** ** **********: . *********
Mflv_1926|M.gilvum_PYR-GCK TADSDPDREWQECLDKAAPIIG---RAVGSLSRA--------
Mvan_4807|M.vanbaalenii_PYR-1 TADSDPDLEWEECLHKAAPIVNAVPVAVPVQPRARSTAS---
MMAR_4488|M.marinum_M TADSDPGAEWEECLHKAAPVVGLPAPTTATRVG---------
MUL_4660|M.ulcerans_Agy99 TADSDPGAEWEECQHKAAPVVGLPAPTTATRVG---------
MLBr_00236|M.leprae_Br4923 TADSDPDAELQECLHKAAPILRLASVATVASVG---------
Mb1032c|M.bovis_AF2122/97 TADSDPDAEWAECLHKAAPIVGLPAATRTTPARLASKVR---
Rv1005c|M.tuberculosis_H37Rv TADSDPDAEWAECLHKAAPIVGLPAATRTTPARLASKVR---
MAV_1127|M.avium_104 TADSDPDAEWAECLHKASPIVGLPSIAAASSAG---------
MSMEG_5446|M.smegmatis_MC2_155 TADSDPDREWEECLHKAASIVGPVTDQSLERSTAS-------
TH_1145|M.thermoresistible__bu TADSDPDREWQECLDKAAPFVGADRGPRLGSRPQDGVVQLTT
MAB_1124c|M.abscessus_ATCC_199 TADSDADAEWQECLHKAAATIGLTAIAELNPAL---------
*****.. * ** .**:. :