For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSLAPVQVIGVLNVTDNSFSDGGRYLDPDDAVQHGLAMVAEGAAIVDVGGESTRPGAIRTDPRVELSRIV PVVKELAAQGITVSIDTTRADVARAALQSGARIVNDVSGGRADPAMAPLVAEAGVAWVLMHWRLMSAERP YEAPNYRDVVAEVRADLLAGVDQAVAAGVDPGSLVIDPGLGFAKTGQHNWALLNALPELVATGVPILLGA SRKRFLGRLLAGADGAVRPPDGRETATAVISALAALHGAWGVRVHDVRASVDALKVVGAWLHAGPQIEKV RCDG
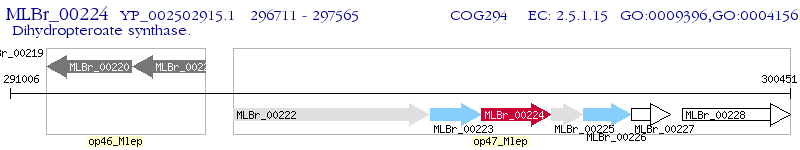
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00224 | folP | - | 100% (284) | dihydropteroate synthase |
| M. leprae Br4923 | MLBr_01063 | folP2 | 2e-33 | 36.43% (280) | dihydropteroate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3638c | folP1 | 1e-121 | 77.11% (284) | dihydropteroate synthase |
| M. gilvum PYR-GCK | Mflv_1418 | - | 1e-110 | 73.96% (265) | dihydropteroate synthase |
| M. tuberculosis H37Rv | Rv3608c | folP1 | 1e-121 | 77.11% (284) | dihydropteroate synthase |
| M. abscessus ATCC 19977 | MAB_0535 | - | 2e-93 | 60.07% (273) | dihydropteroate synthase |
| M. marinum M | MMAR_5111 | folP1 | 1e-121 | 78.39% (273) | dihydropteroate synthase 1 FolP1 |
| M. avium 104 | MAV_0544 | folP | 1e-124 | 84.62% (260) | dihydropteroate synthase |
| M. smegmatis MC2 155 | MSMEG_6103 | folP | 1e-116 | 74.07% (270) | dihydropteroate synthase |
| M. thermoresistible (build 8) | TH_0989 | folP | 1e-110 | 73.00% (263) | DIHYDROPTEROATE SYNTHASE 1 FOLP (DHPS 1) (DIHYDROPTEROATE |
| M. ulcerans Agy99 | MUL_4190 | folP1 | 1e-117 | 77.74% (265) | dihydropteroate synthase 1 FolP1 |
| M. vanbaalenii PYR-1 | Mvan_5370 | - | 1e-107 | 72.90% (262) | dihydropteroate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00224|M.leprae_Br4923 ----------MSLAPVQVIGVLNVTDNSFSDGGRYLDPDDAVQHGLAMVA
MAV_0544|M.avium_104 ---------------------MNVTDDSFSDGGRYLDPDKAVAHGLALVA
Mb3638c|M.bovis_AF2122/97 ----------MSPAPVQVMGVLNVTDDSFSDGGCYLDLDDAVKHGLAMAA
Rv3608c|M.tuberculosis_H37Rv ----------MSPAPVQVMGVLNVTDDSFSDGGCYLDLDDAVKHGLAMAA
MMAR_5111|M.marinum_M ----------MSSRPVQVMGVLNVTNDSFSDGGRYCDEAKAIEHGLALAA
MUL_4190|M.ulcerans_Agy99 ------------------MGVLNVTNDSFSDGGRYCDEAKAIEHGLALAA
Mflv_1418|M.gilvum_PYR-GCK ---------------MQVMGVVNVTDDSFSDGGLFLDRDRAVAHGVELVA
Mvan_5370|M.vanbaalenii_PYR-1 ------------------MGVVNVTDDSFSDGGLFLDRDRAVIHGVELVR
TH_0989|M.thermoresistible__bu VTPLPVLPQTSTTGSTVVMGVVNVTDDSFSDGGLFLDRDRAVAHGLALTE
MSMEG_6103|M.smegmatis_MC2_155 MNPPSLTAQPGTP--VQVMGVVNVTQDSFSDGGKFIDTDRAVEHGLALVA
MAB_0535|M.abscessus_ATCC_1997 ----MIAGVTETPALTHILGVVNVTDDSFSDGGRFVRQSDAVAHGLALAS
:***::****** : *: **: :.
MLBr_00224|M.leprae_Br4923 EGAAIVDVGGESTRPGAIRTDPRVELSRIVPVVKELAAQGITVSIDTTRA
MAV_0544|M.avium_104 EGAGIVDVGGESTRPGATRIDPRVETSRVVPVVKALAAEGVTVSIDTMHA
Mb3638c|M.bovis_AF2122/97 AGAGIVDVGGESSRPGATRVDPAVETSRVIPVVKELAAQGITVSIDTMRA
Rv3608c|M.tuberculosis_H37Rv AGAGIVDVGGESSRPGATRVDPAVETSRVIPVVKELAAQGITVSIDTMRA
MMAR_5111|M.marinum_M EGADIIDVGGESTRPGAARVDPAVEASRVLPVIKELAARGIKVSIDTTRA
MUL_4190|M.ulcerans_Agy99 EGADIIDVGGESTRPGAARVDPAVEASRVLPVIKELAARGIKVSIDTTRA
Mflv_1418|M.gilvum_PYR-GCK QGAAIVDVGGESTRPGATRVDPEVEISRILPVVKELAQQGITVSIDTMHA
Mvan_5370|M.vanbaalenii_PYR-1 QGAAIVDVGGESTRPGATRIDPAVESSRILPVVKELAQLGITVSIDTMHA
TH_0989|M.thermoresistible__bu QGAAVIDVGGESTRPGATRIDPAVEAARVVPVITDLVAQGVTVSIDTMHA
MSMEG_6103|M.smegmatis_MC2_155 AGAQIIDVGGESTRPGATRIDPGVEAARVTPVIRELAAQGITISIDTMHA
MAB_0535|M.abscessus_ATCC_1997 AGADIIDVGGQSTRPGAAPIRQKIEMQRVIPVIKELASHGINISVDTMRA
** ::****:*:**** :* *: **: *. *:.:*:** :*
MLBr_00224|M.leprae_Br4923 DVARAALQSGARIVNDVSGGRADPAMAPLVAEAGVAWVLMHWRLMSAERP
MAV_0544|M.avium_104 DVARAALGSGARIVNDVSGGRADPAMAPLLAEAKVPWVLMHWRSVSAERP
Mb3638c|M.bovis_AF2122/97 DVARAALQNGAQMVNDVSGGRADPAMGPLLAEADVPWVLMHWRAVSADTP
Rv3608c|M.tuberculosis_H37Rv DVARAALQNGAQMVNDVSGGRADPAMGPLLAEADVPWVLMHWRAVSADTP
MMAR_5111|M.marinum_M AVACEALHNGAQIVNDVSGGRADPAMAPLVAEAGVPWVLMHWRPVSDDNP
MUL_4190|M.ulcerans_Agy99 AVACEALHNGAQIVNDVSGGRADPAMAPLVAEASVPWVLMHWRPVSDDNP
Mflv_1418|M.gilvum_PYR-GCK AVAHAALDNGAKIVNDVSGGRADPDMVRVVADAGVPWILMHWRSVDGDNP
Mvan_5370|M.vanbaalenii_PYR-1 AVARVALEHGAQIVNDVSGGRADPDMARVVADAGVPWVLMHWRSVGSDRP
TH_0989|M.thermoresistible__bu DVASAALEHGAHIVNDVSGGRADPNMAKLVAEARVPWVLMHWRSVGATHP
MSMEG_6103|M.smegmatis_MC2_155 DVARAALEAGAHIVNDVSGGRADPGMAGVLAEAKVPWILMHWRSVDADHP
MAB_0535|M.abscessus_ATCC_1997 EVAAAAIDAGANMVNDVSGGRSDPEMAQLIAQRGVPWILMHWRSKDFIHT
** *: **.:********:** * ::*: *.*:***** . .
MLBr_00224|M.leprae_Br4923 YEAPNYRDVVAEVRADLLAGVDQAVAAGVDPGSLVIDPGLGFAKTGQHNW
MAV_0544|M.avium_104 HAAPQYRDVVAEVRAELLASVDAAVAAGVDSARLMIDPGLGFAKTGQHNW
Mb3638c|M.bovis_AF2122/97 HVPVRYGNVVAEVRADLLASVADAVAAGVDPARLVLDPGLGFAKTAQHNW
Rv3608c|M.tuberculosis_H37Rv HVPVRYGNVVAEVRADLLASVADAVAAGVDPARLVLDPGLGFAKTAQHNW
MMAR_5111|M.marinum_M HQVPNYRDVVAEVRAELLTAVDDAIAAGVGPSNLMIDPGLGFAKAGQHNW
MUL_4190|M.ulcerans_Agy99 HQVPNYRDVVAEVRAELLTAVDDAIAAGVGPSNLMIDPGLGFAKAGQHNW
Mflv_1418|M.gilvum_PYR-GCK HEVPAYGDVVAEVRDELLSGVDAAVAAGVHPDKLIIDPGLGFAKTGQHNW
Mvan_5370|M.vanbaalenii_PYR-1 HEVPPYGDVVTAVRDELMAGVDAAVDAGVDPAKLIIDPGLGFAKTAQHNW
TH_0989|M.thermoresistible__bu HRPPRYRDVVAEVRAELLESVDAATSAGVDPANLIIDPGLGFAKTAEHNW
MSMEG_6103|M.smegmatis_MC2_155 HRVPGYRDVVAEVRTELLAAVDAAVTAGVEPERLVIDPGLGFAKTAEHNW
MAB_0535|M.abscessus_ATCC_1997 PATRNYRDVVADVSRELMESVAVAVNAGVSPDKLILDPGLGFAKTAHHNW
* :**: * :*: .* * *** . *::********:..***
MLBr_00224|M.leprae_Br4923 ALLNALPELVATGVPILLGASRKRFLGRLLAGADGAVRPPDGRETATAVI
MAV_0544|M.avium_104 ALLHALPQLVATGIPVLLGASRKRFLGTLLAGPDGAPRPPDGRETATAVI
Mb3638c|M.bovis_AF2122/97 AILHALPELVATGIPVLVGASRKRFLGALLAGPDGVMRPTDGRDTATAVI
Rv3608c|M.tuberculosis_H37Rv AILHALPELVATGIPVLVGASRKRFLGALLAGPDGVMRPTDGRDTATAVI
MMAR_5111|M.marinum_M TLLHALPELVATGIPVLVGASRKRFLGSLLAGPDGAVRPPDGRETATAVI
MUL_4190|M.ulcerans_Agy99 TLLHALPELVATGIPVLVGASRKRFLGSLLAGPDGAVRPPDGRETATAVI
Mflv_1418|M.gilvum_PYR-GCK ALLHALPDFVSTGIPVLVGASRKRFLGTLLAAPDGEPRPPAGRETATAVI
Mvan_5370|M.vanbaalenii_PYR-1 ALLHALPEFVTTGIPVLVGASRKRFLGELLASPDGELRPPAGRETATAVV
TH_0989|M.thermoresistible__bu ALLHALPEFVGTGIPVLVGASRKRFLGALLAGPDGEPRPPDGRETATAVI
MSMEG_6103|M.smegmatis_MC2_155 ALLHALPDLVATGVPVLVGASRKRFLGTLLAAADGTPRPPDGRETATAVI
MAB_0535|M.abscessus_ATCC_1997 LLLQAIPDLQELGYPILIGASRKRFLGSLLTDLKGVERPPDGRETATAVI
:*:*:*:: * *:*:********* **: .* **. **:*****:
MLBr_00224|M.leprae_Br4923 SALAALHGAWGVRVHDVRASVDALKVVGAWLHAGPQIEKVRCDG------
MAV_0544|M.avium_104 SALAALHGAWGVRVHDVRATVDALKVLGAWMGD---------DG------
Mb3638c|M.bovis_AF2122/97 SALAALHGAWGVRVHDVRASVDAIKVVEAWMGAER----IERDG------
Rv3608c|M.tuberculosis_H37Rv SALAALHGAWGVRVHDVRASVDAIKVVEAWMGAER----IERDG------
MMAR_5111|M.marinum_M SALAALHGAWGVRVHDVRASVDALKVVDAWQRAER----TGHDG------
MUL_4190|M.ulcerans_Agy99 SALGALHGAWGVRVHDVRASVDALKVVDAWQRAER----TGRDG------
Mflv_1418|M.gilvum_PYR-GCK SALAGWHDVWGVRVHDVRASVDALAVVAAWRGGAS---------------
Mvan_5370|M.vanbaalenii_PYR-1 SALAGWHHVWGVRVHDVRASVDALAVVAAWQAGRD-------D-------
TH_0989|M.thermoresistible__bu SALAGLAGVWGVRVHDVRASVDALRVMRAWQAGSA-------DG------
MSMEG_6103|M.smegmatis_MC2_155 SVLAAMHGAWGVRVHDVQASVDALKVLGAWTSGGQ-------IG------
MAB_0535|M.abscessus_ATCC_1997 TALGALHEVWGVRVHDVTASMDALKVVRAWETGGAETVEEGEEEAQEAAA
:.*.. .******** *::**: *: **
MLBr_00224|M.leprae_Br4923 --------------------------------------------------
MAV_0544|M.avium_104 --------------------------------------------------
Mb3638c|M.bovis_AF2122/97 --------------------------------------------------
Rv3608c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5111|M.marinum_M --------------------------------------------------
MUL_4190|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1418|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5370|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0989|M.thermoresistible__bu --------------------------------------------------
MSMEG_6103|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0535|M.abscessus_ATCC_1997 PEPPPAPESPPVTIAQDEVTTEVTVRTARPEPSKTAGGRWRREVAQRGAK
MLBr_00224|M.leprae_Br4923 ----
MAV_0544|M.avium_104 ----
Mb3638c|M.bovis_AF2122/97 ----
Rv3608c|M.tuberculosis_H37Rv ----
MMAR_5111|M.marinum_M ----
MUL_4190|M.ulcerans_Agy99 ----
Mflv_1418|M.gilvum_PYR-GCK ----
Mvan_5370|M.vanbaalenii_PYR-1 ----
TH_0989|M.thermoresistible__bu ----
MSMEG_6103|M.smegmatis_MC2_155 ----
MAB_0535|M.abscessus_ATCC_1997 GEPK