For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GVPVYQRILDLFQAEGVNTLFGIPDPNFVHMFLEAESRGWSVVSPHHELSAGFMAEAASRMTGKPGLCIG TLGPGMANIAGAIQCALVENSPVIFLGGQRARVTERRVRRGRIQFVRQEPLFAPSVKYSASIEYADQTDE IIHEALRRAMSGTPGPAYIEYPSHVILSELDVSDPLPPHRYRLVNQTAGEREVAEAVRLIRAAESPILLV GHGVHTSRTGAAVRELAELMACPVIQTSGGTSFIDGLQDRTFAYGFSPAAVEAVVTSDLCLALGTELGEP SHYGRTRHWAQNDATRKWILVEQDPAAIGVNRPIDVPLVGDLRGVVPQLVAALRDTPRKPSPDLDRWIEQ DAAELAQLAENAPTGRTPIHPARFVVEATRAFPDDGIMVRDGGATVIFGWTYSQAKPRDVIWNQNFGHLG TGLPYAVGASIAEGGKRPVMLLTSDSAFLFHIAELETAARQGLPLVCVVGVDHQWGLEVGVYKRTFSQPS PQPGVHWSKDVRLDKVAEGFGCHGEYVEKEDEIGPAIQRSYASGKPAVVHVCIDPKANSEEMPKYDEFRT WYAEGTQ*
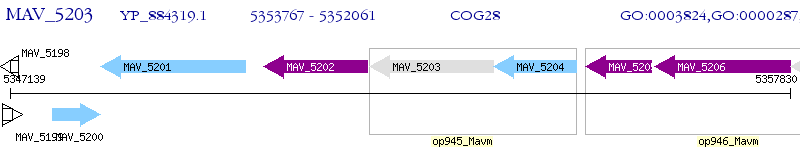
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5203 | - | - | 100% (568) | thiamine pyrophosphate binding domain-containing protein |
| M. avium 104 | MAV_2895 | - | 2e-48 | 28.21% (546) | hypothetical protein MAV_2895 |
| M. avium 104 | MAV_4935 | - | 7e-38 | 24.73% (562) | acetolactate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1851 | ilvG | 5e-45 | 27.68% (542) | hypothetical protein Mb1851 |
| M. gilvum PYR-GCK | Mflv_1796 | - | 0.0 | 86.34% (571) | thiamine pyrophosphate enzyme, central region |
| M. tuberculosis H37Rv | Rv1820 | ilvG | 5e-45 | 27.68% (542) | hypothetical protein Rv1820 |
| M. leprae Br4923 | MLBr_02083 | ilvG | 3e-47 | 27.46% (539) | hypothetical protein MLBr_02083 |
| M. abscessus ATCC 19977 | MAB_3323c | - | 6e-39 | 27.82% (568) | acetolactate synthase 1 catalytic subunit |
| M. marinum M | MMAR_2697 | ilvG | 5e-47 | 28.83% (548) | acetolactate synthase IlvG |
| M. smegmatis MC2 155 | MSMEG_5297 | - | 2e-44 | 27.96% (558) | putative oxalyl-CoA decarboxylase |
| M. thermoresistible (build 8) | TH_4403 | - | 0.0 | 83.98% (568) | PUTATIVE thiamine pyrophosphate enzyme, central domain family |
| M. ulcerans Agy99 | MUL_3039 | ilvG | 1e-45 | 28.88% (547) | hypothetical protein MUL_3039 |
| M. vanbaalenii PYR-1 | Mvan_4954 | - | 0.0 | 84.41% (571) | thiamine pyrophosphate binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1796|M.gilvum_PYR-GCK ------------------------MGVP-------VYKRILDLFEAEGVN
Mvan_4954|M.vanbaalenii_PYR-1 ----------------------MTMGIP-------VYKRILDLFEAEGVN
MAV_5203|M.avium_104 ------------------------MGVP-------VYQRILDLFQAEGVN
TH_4403|M.thermoresistible__bu ------------------------MGVP-------VYKRILDLFEAEGIN
MMAR_2697|M.marinum_M ----------------MSTDAAPVQTVH-------AGRLVARRLRASGVD
MUL_3039|M.ulcerans_Agy99 ----------------MSTDATPVQTVH-------AGRLVARRLRASGVD
Mb1851|M.bovis_AF2122/97 ----------------MSTDTAPAQTMH-------AGRLIARRLKASGID
Rv1820|M.tuberculosis_H37Rv ----------------MSTDTAPAQTMH-------AGRLIARRLKASGID
MLBr_02083|M.leprae_Br4923 ----------------MSTDPVLTHSIH-------AGRLVVRRLKASGVD
MSMEG_5297|M.smegmatis_MC2_155 ----------------MNTAPLMTESVPGTSPVTDGAHVLVDALKLNGVE
MAB_3323c|M.abscessus_ATCC_199 MSAPAKPKPASASGRAPGTAPEPNTVAPQR---VTGAQSVVRSLEELGVE
: : :. *::
Mflv_1796|M.gilvum_PYR-GCK TLFGIPDPNFVHMFTEADA-RGWSVVAPHHELSAGFMAEAASRMTGGPGL
Mvan_4954|M.vanbaalenii_PYR-1 TLFGIPDPNFVHMFTEADA-RGWSVVAPHHELSAGFMAEAASRMTGNPGL
MAV_5203|M.avium_104 TLFGIPDPNFVHMFLEAES-RGWSVVSPHHELSAGFMAEAASRMTGKPGL
TH_4403|M.thermoresistible__bu TLFGIPDPNFVHLFLEAET-RGWSVVAPHHEESAGFMAEAASRMTGRPGL
MMAR_2697|M.marinum_M TVFTLSGGHLFSLYDGCRD-EGVRLIDTRHEQAAAFAAEGWSKVTRVPGV
MUL_3039|M.ulcerans_Agy99 TVFTLSGGHLFSLYDGCRD-EGVRLVDTRHEQTAAFAAEGWSKVTRVPGV
Mb1851|M.bovis_AF2122/97 TVFTLSGGHLFSIYDGCRE-EGIRLIDTRHEQTAAFAAEGWSKVTRVPGV
Rv1820|M.tuberculosis_H37Rv TVFTLSGGHLFSIYDGCRE-EGIRLIDTRHEQTAAFAAEGWSKVTRVPGV
MLBr_02083|M.leprae_Br4923 TVFTLSGGHLFSIYDGCRS-ESIRLIDTRHEQTAAFAAEGWSKVTRQPGV
MSMEG_5297|M.smegmatis_MC2_155 TLYGVVGIPITDVARVAQA-QGLRYIGFRHESDAGHAAAAAGFLTKKPGF
MAB_3323c|M.abscessus_ATCC_199 VIFGIPGGAVLPVYDPLYDSTKVRHVLVRHEQGAGHAASGYAHANGKVGV
.:: : . . : : :** *.. * . . . *.
Mflv_1796|M.gilvum_PYR-GCK CIGTLGPGVANIAGAMMCALVENSPVIFLGGQRARVTERRVRRGRIQFVQ
Mvan_4954|M.vanbaalenii_PYR-1 CIGTLGPGVANIAGAMMCALVENSPVIFLGGQRARITERRVRRGRIQFVQ
MAV_5203|M.avium_104 CIGTLGPGMANIAGAIQCALVENSPVIFLGGQRARVTERRVRRGRIQFVR
TH_4403|M.thermoresistible__bu CIGTLGPGLANIAGAMMCAKVENSPVIFLGGQRARITERRVRRGRIQFVQ
MMAR_2697|M.marinum_M AALTAGPGVTNGMSAMAAAQQNQSPLVVLGG---RAPAGRWGMGSLQEID
MUL_3039|M.ulcerans_Agy99 AALTAGPGVTNGMSAMAAAQQNQSPLVVLGG---RAPAGRWGMGSLQEID
Mb1851|M.bovis_AF2122/97 AALTAGPGITNGMSAMAAAQQNQSPLVVLGG---RAPALRWGMGSLQEID
Rv1820|M.tuberculosis_H37Rv AALTAGPGITNGMSAMAAAQQNQSPLVVLGG---RAPALRWGMGSLQEID
MLBr_02083|M.leprae_Br4923 AVLTAGPGVTNGMSAMAAAQQNQSPLVVLGG---RAPALRWGMGSLQEID
MSMEG_5297|M.smegmatis_MC2_155 CLTVSAPGFLNGLVALANATTNCFPMVQISGS-SDRHIVDLQRGDYEEMD
MAB_3323c|M.abscessus_ATCC_199 MMATSGPGATNLVTPLADAQMDSIPVVAITG---QVGRGLIGTDAFQEAD
. .** * .: * : *:: : * . :
Mflv_1796|M.gilvum_PYR-GCK QEGLFAPSVKYSSSIEYADQTDEIIREAIRRSMSGTPGPSYVEFPSHVIL
Mvan_4954|M.vanbaalenii_PYR-1 QEGLFAPSVKYSSSIEYADQTDEIVHEAIRRAMSGTPGPAYIEFPSHVIL
MAV_5203|M.avium_104 QEPLFAPSVKYSASIEYADQTDEIIHEALRRAMSGTPGPAYIEYPSHVIL
TH_4403|M.thermoresistible__bu QEGMLAPMVKFSSSIEYAEQTDEIIREAIRRAMSGTPGPAYVEYPQHVIL
MMAR_2697|M.marinum_M HVPFVAPLARFAATAQSAEDAGRLVDQALRAAVGSRSGVAFVDFPMDHVF
MUL_3039|M.ulcerans_Agy99 HVPFVAPLTRFAATAQSAEDAGLLVDQALRAAVGSHSGVAFVDFPMDHVF
Mb1851|M.bovis_AF2122/97 HVPFVAPVARFAATAQSAENAGLLVDQALQAAVSAPSGVAFVDFPMDHAF
Rv1820|M.tuberculosis_H37Rv HVPFVAPVARFAATAQSAENAGLLVDQALQAAVSAPSGVAFVDFPMDHAF
MLBr_02083|M.leprae_Br4923 HVPFVAPLARFAATAQSADAAGRLVDEALQAAVVAPSGVGFVDFPMDYVF
MSMEG_5297|M.smegmatis_MC2_155 QLAAAKPFVKAAFRVNRAADIGLAVARAIRTASSGRPGGVYLDIPAAVLG
MAB_3323c|M.abscessus_ATCC_199 ISGITMPITKHNFLVSNAEDIPKVIAEAFYIAQSGRPGAVLVDIPKDILQ
* .: . * : .*: : . .* :: *
Mflv_1796|M.gilvum_PYR-GCK EELDVADPLPPERYRLVNQG----AGEREIAEAARLIRAAERPILLVGHG
Mvan_4954|M.vanbaalenii_PYR-1 DELNVGEAPEPSTYRLLNQG----AGAREVAEAVELIRQAERPILLVGHG
MAV_5203|M.avium_104 SELDVSDPLPPHRYRLVNQT----AGEREVAEAVRLIRAAESPILLVGHG
TH_4403|M.thermoresistible__bu EELDVPDPLPPHRYRLVDQR----AGAAEIAEAVRLIRAAEHPILLVGHA
MMAR_2697|M.marinum_M SMAEDDGR-PGVLTEQPAAAQHPAPNGREIDRAAELLSRAHRPVIMAGTN
MUL_3039|M.ulcerans_Agy99 SMAQDDGR-PGVLTEQPAAAQHPAPNGREIDRAAELLSRAQRPVIMAGTN
Mb1851|M.bovis_AF2122/97 SMSSDNGR-PGALTELPAG---PTPAGDALDRAAGLLSTAQRPVIMAGTN
Rv1820|M.tuberculosis_H37Rv SMSSDNGR-PGALTELPAG---PTPAGDALDRAAGLLSTAQRPVIMAGTN
MLBr_02083|M.leprae_Br4923 SMADDVGGGPGALANLPVA---PAANPEALDRAAGLLAAAHRPVIMAGTN
MSMEG_5297|M.smegmatis_MC2_155 EVIDAGAAAASLRTVVDAAP-EQLPPPRAVDRAVALLAAARRPLVVLGKG
MAB_3323c|M.abscessus_ATCC_199 AQTTFSWPPQIYLPGYKPVT---KPHGKQVREAVRLIAAAKRPVLYVGGG
. : .*. *: *. *:: *
Mflv_1796|M.gilvum_PYR-GCK VHTSRTQQQVRELAELMACPVIQTSGGTSFIPGLQDRTFPYLFSP-----
Mvan_4954|M.vanbaalenii_PYR-1 VHTSRTQEQVRELAELMACPVIQTSGGTSFIPGLEDRTFPYLFSP-----
MAV_5203|M.avium_104 VHTSRTGAAVRELAELMACPVIQTSGGTSFIDGLQDRTFAYGFSP-----
TH_4403|M.thermoresistible__bu VHTSRTQQPVRELAELMGCPVIQTSGGTSFIPGIEDRTFPYGFSP-----
MMAR_2697|M.marinum_M VWWGHGERALLRLAEERQIPVLMNG----MARGVVPADHPLAFSR-----
MUL_3039|M.ulcerans_Agy99 VWWGHGERALLRLAEERQIPVLMNG----MARGVVPADHPLAFSR-----
Mb1851|M.bovis_AF2122/97 VWWGHAEAALLRLVEERHIPVLMNG----MARGVVPADHRLAFSR-----
Rv1820|M.tuberculosis_H37Rv VWWGHAEAALLRLVEERHIPVLMNG----MARGVVPADHRLAFSR-----
MLBr_02083|M.leprae_Br4923 VWWGHAEAALLRLAEECQIPVLMNG----MARGVVPADHPLAFSR-----
MSMEG_5297|M.smegmatis_MC2_155 AAYAQADKTIRNFLESTGLPFLPMS----MAKGLLPDTHPQSVAS-----
MAB_3323c|M.abscessus_ATCC_199 VIRADASPELKELAELTGIPVVTTL----MARGAFPDSHELHMGMPGMHG
. . : .: * *.: : * . ..
Mflv_1796|M.gilvum_PYR-GCK --AANEAVEQSDLCVALGTELGEPMHYGRTQHWAGNDANRKWILVEQDPT
Mvan_4954|M.vanbaalenii_PYR-1 --AANEAVEQSDLCVALGTELGEPMHYGRTQHWAGNDANRKWVYVEQDPT
MAV_5203|M.avium_104 --AAVEAVVTSDLCLALGTELGEPSHYGRTRHWAQNDATRKWILVEQDPA
TH_4403|M.thermoresistible__bu --AAIEAVRTSDLCVALGTELGEPVHFGRHRHWVDNEANRKWVYVEQDPA
MMAR_2697|M.marinum_M --ARSKALGEADVALIVGVPMDFRLGFG-----GVFGPQTQLIVVDRAEP
MUL_3039|M.ulcerans_Agy99 --ARSKALGEADVALIVGVPMDFRLGFG-----GVFGPQTQLIVVDRAEP
Mb1851|M.bovis_AF2122/97 --ARSKALGEADVALIVGVPMDFRLGFG-----GVFGSTTQLIVADRVEP
Rv1820|M.tuberculosis_H37Rv --ARSKALGEADVALIVGVPMDFRLGFG-----GVFGSTTQLIVADRVEP
MLBr_02083|M.leprae_Br4923 --ARGKALGEADVALVVGVPMDFRLGFG-----KVFGSQTQLVVADRAEP
MSMEG_5297|M.smegmatis_MC2_155 --ARSMALRDADVVLLVGARLNWLLGHGEP---PQFSRDVKFIQVDIDPG
MAB_3323c|M.abscessus_ATCC_199 TVGAVAALQRSDLLITLGARFDDRVTGQ----LDSFAPEAKVIHADIDPA
. *: :*: : :*. :. : : .:
Mflv_1796|M.gilvum_PYR-GCK AIGVNRPIDVPLVGDLRGVVPQLVDALRDTPRA--PSANLDSLVKADAAE
Mvan_4954|M.vanbaalenii_PYR-1 AIGVNRSFDVPLVGDLRGVVPQLVEALRDTTRA--PSANLETLVDADAAE
MAV_5203|M.avium_104 AIGVNRPIDVPLVGDLRGVVPQLVAALRDTPRK--PSPDLDRWIEQDAAE
TH_4403|M.thermoresistible__bu AIGVNRPIDVPLIGDLRGVVPQLVEALREQPRQ--PSPDLDRWIRQDAER
MMAR_2697|M.marinum_M ERQHPRPIAAGLYGDLSQILAALTGPDAGDHQ-----SWIRELRDTETEA
MUL_3039|M.ulcerans_Agy99 ERQHPRPIAAGLYGDLSQILAALTGPDAGDHQ-----SWIRELRDTETEA
Mb1851|M.bovis_AF2122/97 AREHPRPVAAGLYGDLTATLSALAGSGGTDHQ-----GWIEELATAETMA
Rv1820|M.tuberculosis_H37Rv AREHPRPVAAGLYGDLTATLSALAGSGGTDHQ-----GWIEELATAETMA
MLBr_02083|M.leprae_Br4923 DRDYPRPVAAGLYGDLPAILSALAGVGGTDSQ-----DWIRALRTAETAT
MSMEG_5297|M.smegmatis_MC2_155 ELDSNAPIAAPLVGDIGSVIEALAERAQFIDAP---APWREQLAERKARN
MAB_3323c|M.abscessus_ATCC_199 EIGKNRHADVPIVGDVKNVIIELIEAIRADRAAGTVADPLSEWWEYLSDV
. : **: : * :
Mflv_1796|M.gilvum_PYR-GCK LARVAEEAPSGRAPVHPARYVVEATKAFNEHTEDGILVRDGGATVIFQWT
Mvan_4954|M.vanbaalenii_PYR-1 LARVAEEAPSGRTPVHPARFVVEATKAFNDYAQDGILVRDGGATVIFQWT
MAV_5203|M.avium_104 LAQLAENAPTGRTPIHPARFVVEATRAFPD---DGIMVRDGGATVIFGWT
TH_4403|M.thermoresistible__bu LAELAETAPTGRKPVHPARFVVEATKALPD---DAILVRDGGATVIFQWT
MMAR_2697|M.marinum_M REREKTELAEDRIPLHPMRVYAELAPLLDR---DAIVVIDAGDFGSYAGR
MUL_3039|M.ulcerans_Agy99 REREKTELAEDRIPLHPMRVYAELAPLLDR---DAIVVIDAGDFGSYAGR
Mb1851|M.bovis_AF2122/97 RDLEKAELVDDRIPLHPMRVYAELAALLER---DALVVIDAGDFGSYAGR
Rv1820|M.tuberculosis_H37Rv RDLEKAELVDDRIPLHPMRVYAELAALLER---DALVVIDAGDFGSYAGR
MLBr_02083|M.leprae_Br4923 RDLEKAELDDDRLPLHPMRVYAELASLLDR---DAIVVIDAGDFGSYAGR
MSMEG_5297|M.smegmatis_MC2_155 AESMAKRLAADPRPMRFHGALRAIRDVLRDRP-DVYVVNEGANALDLARN
MAB_3323c|M.abscessus_ATCC_199 RKTYPLGYDAQHDGSLGPEFVIETLGKIAG--PDAVYVAGVGQHQMWAAQ
: * * .
Mflv_1796|M.gilvum_PYR-GCK YSQVKPRDVIWNQN-FGHLGTGLPYAVGASVAEGRKRPVMLLTSDSAFLF
Mvan_4954|M.vanbaalenii_PYR-1 YSQAKPRDVIWNQN-FGHLGTGLPYAVGASVAEGRTRPVMLLTSDSAFLF
MAV_5203|M.avium_104 YSQAKPRDVIWNQN-FGHLGTGLPYAVGASIAEGGKRPVMLLTSDSAFLF
TH_4403|M.thermoresistible__bu YSQAKPRDVIWNQN-FGHLGTGLPYAVGASVAEGGKRPVMLLTSDSAFLF
MMAR_2697|M.marinum_M VIDSYQPGCWLDSGPFGCLGSGPGYALAAKLARP-ERQVVLLQGDGAFGF
MUL_3039|M.ulcerans_Agy99 VIDSYQPGCWLDSGPFGCLGSGPGCALAAKLARP-ERQVVLLQGDGAFGF
Mb1851|M.bovis_AF2122/97 MIDSYLPGCWLDSGPFGCLGSGPGYALAAKLARP-QRQVVLLQGDGAFGF
Rv1820|M.tuberculosis_H37Rv MIDSYLPGCWLDSGPFGCLGSGPGYALAAKLARP-QRQVVLLQGDGAFGF
MLBr_02083|M.leprae_Br4923 VIDSYLPGCWLDSGPFGCLGSGPGYALAAKLARP-NRQVVLLQGDGAFGF
MSMEG_5297|M.smegmatis_MC2_155 VIDMAVPRHRLDSGTWGVMGVGMGYAIAAAVEGS--GPVVAIEGDSAFGF
MAB_3323c|M.abscessus_ATCC_199 FIKYENPRTWLNSGGLGTMGFAVPAAMGAKMARP-EAEVWAIDGDGCFQM
. :.. * :* . *:.* : * : .*..* :
Mflv_1796|M.gilvum_PYR-GCK HIAELETAVRENLPLVCVVGVDHQWGLEVGVYKRTFEQ--PSPQPGVHWS
Mvan_4954|M.vanbaalenii_PYR-1 HIAELETAARENVPLVCVVGVDHQWGLEVGVYKRTFEQ--PSPQPGVHWS
MAV_5203|M.avium_104 HIAELETAARQGLPLVCVVGVDHQWGLEVGVYKRTFSQ--PSPQPGVHWS
TH_4403|M.thermoresistible__bu HIAELETAARQNLPLVCVVGVDHAWGLEVGVYKRTFEQ--PSPQPGVHWS
MMAR_2697|M.marinum_M SGMEWDTLVRHNVPVVSVVGNNGIWGLEKHPMEALYG-----YSVVAELR
MUL_3039|M.ulcerans_Agy99 SGMEWDTLVRHNVPVVSVVGNNGIWGLEKHPMEALYG-----YSVVAELR
Mb1851|M.bovis_AF2122/97 SGMEWDTLVRHNVAVVSVIGNNGIWGLEKHPMEALYG-----YSVVAELR
Rv1820|M.tuberculosis_H37Rv SGMEWDTLVRHNVAVVSVIGNNGIWGLEKHPMEALYG-----YSVVAELR
MLBr_02083|M.leprae_Br4923 SGMEWDTLVRHNVPVVSVIGNNGIWGLEKHPMEALYG-----YSVVAELR
MSMEG_5297|M.smegmatis_MC2_155 SAMELETICRYRLPVVVVVLNNGGVYRGDGHNPASDD------PSPTTLM
MAB_3323c|M.abscessus_ATCC_199 TNQELATCAIEGVPIKVALINNGNLGMVRQWQTLFYEERYSQTDLATHSR
* * :.: .: : .
Mflv_1796|M.gilvum_PYR-GCK KDVRMDKIAEGFGCHGEYVEKEDEIGPAIQRAFASG-KTAVVHVCIDPKA
Mvan_4954|M.vanbaalenii_PYR-1 KDVRMDKIAEGFGCHGEYVEKEDEIGPAIQRAYASG-RTAVVHVCIDPKA
MAV_5203|M.avium_104 KDVRLDKVAEGFGCHGEYVEKEDEIGPAIQRSYASG-KPAVVHVCIDPKA
TH_4403|M.thermoresistible__bu KNVRFDKIAEGLDCHGEFVDSEEEIGPAIKRAYASG-RTAVVHVAIDPKA
MMAR_2697|M.marinum_M PGTRYDEVVRALGGHGELVAAPAQLRPALERAFASG-LPAVVNVLTDPSV
MUL_3039|M.ulcerans_Agy99 PGTRYDEVVRALGGHGELVAAPAELRPALERAFASG-LPAVVNVLTDSSV
Mb1851|M.bovis_AF2122/97 PGTRYDEVVRALGGHGELVSVPAELRPALERAFASG-LPAVVNVLTDPSV
Rv1820|M.tuberculosis_H37Rv PGTRYDEVVRALGGHGELVSVPAELRPALERAFASG-LPAVVNVLTDPSV
MLBr_02083|M.leprae_Br4923 PGTRYDEVVRALGGHGELVSVPGELRPALERAFVTG-LPSVVNVLTDPSV
MSMEG_5297|M.smegmatis_MC2_155 PAARHDRLIEAFGGTGHHVTTPAELGAALTEALASG-GPALIDCVIDPAD
MAB_3323c|M.abscessus_ATCC_199 RIPDFVKLAEALGCVGLRCEREEDVAKVIEEARAINDRPVVIDFIVGADA
.: ..:. * :: .: .: . . . ::. ..
Mflv_1796|M.gilvum_PYR-GCK NSEE--MPKYDRFRTWYAEGTQ------------
Mvan_4954|M.vanbaalenii_PYR-1 NSEE--MPKYDRFRTWYAEGTQ------------
MAV_5203|M.avium_104 NSEE--MPKYDEFRTWYAEGTQ------------
TH_4403|M.thermoresistible__bu NSEE--MPSYDEFRTWYAEGQQ------------
MMAR_2697|M.marinum_M AYPR--RSNLA-----------------------
MUL_3039|M.ulcerans_Agy99 AYPR--RSNLA-----------------------
Mb1851|M.bovis_AF2122/97 AYPR--RSNLA-----------------------
Rv1820|M.tuberculosis_H37Rv AYPR--RSNLA-----------------------
MLBr_02083|M.leprae_Br4923 AYPR--RSNLA-----------------------
MSMEG_5297|M.smegmatis_MC2_155 GTESGHLTQLNPAVVGHHPATN------------
MAB_3323c|M.abscessus_ATCC_199 QVWPMVAAGTSNDEIQAARGIRPLFDDDELVADE
.