For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSVPARRGDAHIDFARSPRPTIGVEWEFALVDAQTRDLSNEATAVIAEIGENPRVHKELLRNTVEVVSGI CRTVPEAMEDLRQTLGPARRIVRDRGMELFCAGAHPFAQWTTQKLTDAPRYAELIKRTQWWGRQMLIWGV HVHVGISSPNKVMPIMTSLLNYYPHLLALSASSPWWTGVDTGYASNRAMMFQQLPTAGLPFQFQTWAEFE GFVYDQKKTGIIDHVDEVRWDIRPSPHLGTLEMRICDGVSNLHELAALVALTHCLVVDLDRRLEADESLP TMPPWHHQENKWRAARYGLDAVIILDADSNERLVTEDLDDVLNRLEPVARKLQCADELAAVADIPRHGAS YQRQRRVAEEHDGDLRAVVDALVAELEI*
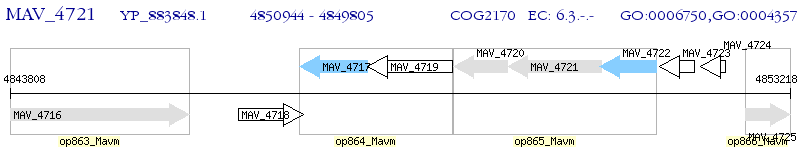
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4721 | - | - | 100% (379) | carboxylate-amine ligase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0441 | - | 0.0 | 86.97% (376) | carboxylate-amine ligase |
| M. gilvum PYR-GCK | Mflv_0171 | - | 0.0 | 81.65% (376) | carboxylate-amine ligase |
| M. tuberculosis H37Rv | Rv0433 | - | 0.0 | 86.97% (376) | carboxylate-amine ligase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4183c | - | 1e-171 | 78.96% (366) | carboxylate-amine ligase |
| M. marinum M | MMAR_0748 | - | 0.0 | 86.42% (383) | hypothetical protein MMAR_0748 |
| M. smegmatis MC2 155 | MSMEG_0836 | - | 1e-179 | 80.47% (379) | carboxylate-amine ligase |
| M. thermoresistible (build 8) | TH_0948 | - | 0.0 | 82.18% (376) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1380 | - | 0.0 | 86.16% (383) | carboxylate-amine ligase |
| M. vanbaalenii PYR-1 | Mvan_0736 | - | 0.0 | 80.42% (378) | carboxylate-amine ligase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0748|M.marinum_M ---------MSSAPASSSAGRTANRIDFARSPRPTVGVEWEFALVDAETR
MUL_1380|M.ulcerans_Agy99 MMPVSGWRAVSSAPASSSAGRTANRIDFARSPRPTVGVEWEFALVDAETR
MAV_4721|M.avium_104 ---------MSSVPAR----RGDAHIDFARSPRPTIGVEWEFALVDAQTR
Mb0441|M.bovis_AF2122/97 ------------MPAR----RSAARIDFAGSPRPTLGVEWEFALVDSQTR
Rv0433|M.tuberculosis_H37Rv ------------MPAR----RSAARIDFAGSPRPTLGVEWEFALVDSQTR
MSMEG_0836|M.smegmatis_MC2_155 ---------MSSLPAD-------SRIDFAGSPRPTVGVEWEFALVDAKTR
TH_0948|M.thermoresistible__bu ---------VSSPAGS-------TRIDFAGSPRPTLGVEWEFALVDADTR
Mflv_0171|M.gilvum_PYR-GCK ---------MSSPVPDQ---RPDSRIDFNGSVRPTLGVEWEFALVDAQTR
Mvan_0736|M.vanbaalenii_PYR-1 ---------MSSLAADQ---RADGRIDFVGSVRPTVGVEWEFALVDATTR
MAB_4183c|M.abscessus_ATCC_199 ---------------------MARPIEFRGSPRPTLGVEWEFALVDAQTR
*:* * ***:**********: **
MMAR_0748|M.marinum_M DLSNEATAVIAEIGENPRVHKELLRNTVEVVSGVCGSAGEAIEDLRTTLG
MUL_1380|M.ulcerans_Agy99 DLSNEATAVIAEIGENPRVHKELLRNTVEVVSGVCGSAGEAIEDLRTTLG
MAV_4721|M.avium_104 DLSNEATAVIAEIGENPRVHKELLRNTVEVVSGICRTVPEAMEDLRQTLG
Mb0441|M.bovis_AF2122/97 DLSNEATAVIAEIGENPRVHKELLRNTVEIVSGICECTAEAMQDLRDTLG
Rv0433|M.tuberculosis_H37Rv DLSNEATAVIAEIGENPRVHKELLRNTVEIVSGICECTAEAMQDLRDTLG
MSMEG_0836|M.smegmatis_MC2_155 ELSNEAAAVIAEIGENPHVHKELLRNTVEIVTGICENSGEAMGDLCDTLS
TH_0948|M.thermoresistible__bu DLSNEAAEVIAEIGENPHVHKELLRNTVEFVTGICADTPEAIADLRATLL
Mflv_0171|M.gilvum_PYR-GCK DLSNEAASVIAEIGENPRVHKELLRNTVEVVTGICDNSGQAMDDLASTLR
Mvan_0736|M.vanbaalenii_PYR-1 DLSNEAASVIAEIGENPRVHKELLRNTVEVVTGICANAGEAMEDLASTLR
MAB_4183c|M.abscessus_ATCC_199 DLTNAASAVLAAIGETPHVHKELLRNTIEVVTGICGTVGEAMHDLQTTLA
:*:* *: *:* ***.*:*********:*.*:*:* :*: ** **
MMAR_0748|M.marinum_M PARRIVHARGMELFCAGTHPFAQWSTQKLTDAPRYAELIKRTQWWGRQML
MUL_1380|M.ulcerans_Agy99 PARRIVHARGMELFCEGTHPFAQWSTQKLTDAPRYAELIKRTQWWGRQML
MAV_4721|M.avium_104 PARRIVRDRGMELFCAGAHPFAQWTTQKLTDAPRYAELIKRTQWWGRQML
Mb0441|M.bovis_AF2122/97 PARQIVRDRGMELFCAGTHPFARWSAQKLTDAPRYAELIKRTQWWGRQML
Rv0433|M.tuberculosis_H37Rv PARQIVRDRGMELFCAGTHPFARWSAQKLTDAPRYAELIKRTQWWGRQML
MSMEG_0836|M.smegmatis_MC2_155 TVRRAVRGRGMELFCAGTHPFGKWSAAQLTDAPRYAELIKRTQWWGRQML
TH_0948|M.thermoresistible__bu PARKIVRDRGMELFCAGTHPFAKWSAQKLTDAPRYAELIKRTQWWGRQML
Mflv_0171|M.gilvum_PYR-GCK PVREIVRERGMELFCAGTHPFADWSAQKLTDAPRYAELIKRTQWWGRQMT
Mvan_0736|M.vanbaalenii_PYR-1 PVREVVRERGMDLFCAGTHPFADWSVQKLTDAPRYAELIKRTQWWGRQML
MAB_4183c|M.abscessus_ATCC_199 TVRPIVQSMGMELFCAGTHPFARWSAQQLTDAPRYDELIARTQWWGRQML
..* *: **:*** *:***. *:. :******* *** *********
MMAR_0748|M.marinum_M IWGVHVHVGVSSANKVMPIISALLNYYPHLLALSASSPWWGGEDTGYASN
MUL_1380|M.ulcerans_Agy99 IWGVHVHVGVSSANKVMPIISALLNYYPHLLALSASSPWWGGEDTGYASN
MAV_4721|M.avium_104 IWGVHVHVGISSPNKVMPIMTSLLNYYPHLLALSASSPWWTGVDTGYASN
Mb0441|M.bovis_AF2122/97 IWGVHVHVGIRSAHKVMPIMTSLLNYYPHLLALSASSPWWGGEDTGYASN
Rv0433|M.tuberculosis_H37Rv IWGVHVHVGIRSAHKVMPIMTSLLNYYPHLLALSASSPWWGGEDTGYASN
MSMEG_0836|M.smegmatis_MC2_155 IWGVHVHVGVSSAHKVMPIISSLLNQYPHLLALSASSPFWDGEDTGYASN
TH_0948|M.thermoresistible__bu IWGVHVHVGVSSAHKVMPIISSLLNQYPHLLALSASSPYWDGEDTGYASN
Mflv_0171|M.gilvum_PYR-GCK IWGVHVHVGVSSAHKVMPIITSLLHQYPHLLALSASSPFWDGEDTGYASN
Mvan_0736|M.vanbaalenii_PYR-1 IWGVHVHVGVSSAHKVMPIITALLHQYPHLLALSASSPYWDGEDTGYASN
MAB_4183c|M.abscessus_ATCC_199 IWGVHVHVGISSKHKVMPIVTSMLNYYPHLLALSASSPMWEGQDTGYASN
*********: * :*****::::*: ************ * * *******
MMAR_0748|M.marinum_M RAMMFQQLPTAGLPFQFQTWSEFEGFVYDQKKTGIIDHINEIRWDIRPSP
MUL_1380|M.ulcerans_Agy99 RAMMFQQLPTAGLPFQFQTWSEFEGFVYDQKKTGIIDHINEIRWDIRPSP
MAV_4721|M.avium_104 RAMMFQQLPTAGLPFQFQTWAEFEGFVYDQKKTGIIDHVDEVRWDIRPSP
Mb0441|M.bovis_AF2122/97 RAMMFQQLPTAGLPFHFQRWAEFEGFVYDQKKTGIIDHMDEIRWDIRPSP
Rv0433|M.tuberculosis_H37Rv RAMMFQQLPTAGLPFHFQRWAEFEGFVYDQKKTGIIDHMDEIRWDIRPSP
MSMEG_0836|M.smegmatis_MC2_155 RAMMFQQLPTAGLPFQFQTWHEFEGFVHDQKKTGIIDHLSEIRWDIRPSP
TH_0948|M.thermoresistible__bu RAMMFQQLPTAGLPFQFQTWAEFERFVADQKKTGIIDHMNEIRWDIRPSP
Mflv_0171|M.gilvum_PYR-GCK RAMMFQQLPTAGLPFHFQEWREWERFVSDQKKTGIIDHMNEIRWDIRPSP
Mvan_0736|M.vanbaalenii_PYR-1 RAMMFQQLPTAGLPFHFQEWREFERFVSDQKKTGIIDHMNEIRWDIRPSP
MAB_4183c|M.abscessus_ATCC_199 RAMMFQQLPTAGLPFHFQAWEQFEGFVDDQMKTGIIDQLNEIRWDIRPSP
***************:** * ::* ** ** ******::.*:********
MMAR_0748|M.marinum_M HLGTIELRICDGVSNLRELGALVALMHCLVVDLDRRLDAGETLPTMPPWH
MUL_1380|M.ulcerans_Agy99 HLGTIELRICDGVSNLRELGALVALMHCLVVDLDRRLDAGETLPTMPPWH
MAV_4721|M.avium_104 HLGTLEMRICDGVSNLHELAALVALTHCLVVDLDRRLEADESLPTMPPWH
Mb0441|M.bovis_AF2122/97 HLGTLEVRICDGVSNLRELGALVALTHCLIVDLDRRLDAGETLPTMPPWH
Rv0433|M.tuberculosis_H37Rv HLGTLEVRICDGVSNLRELGALVALTHCLIVDLDRRLDAGETLPTMPPWH
MSMEG_0836|M.smegmatis_MC2_155 QLGTVEVRIFDGVSNVRELSALVALTHCLIVDLDRRLDAGEQLPVMPPWH
TH_0948|M.thermoresistible__bu HLGTIEVRVFDGISNLRELGALVALTHCLVVDLDRRLDAGEQLPTMPPWH
Mflv_0171|M.gilvum_PYR-GCK HLGTIEVRIFDGVSNLHELSALVALTHCLIVDLDRRIEAGESLPVMPPWH
Mvan_0736|M.vanbaalenii_PYR-1 HLGTIEVRIFDGVSNLHELSALVALTHCLIVDLDRRLDAGESLPVMPPWH
MAB_4183c|M.abscessus_ATCC_199 ALGTVEVRIFDGISNVAELEALVALTHCLVVDLDRKLEAGESLPTMPPWH
***:*:*: **:**: ** ***** ***:*****:::*.* **.*****
MMAR_0748|M.marinum_M VQENKWRAARYGLDAVIILDAESNERLVTDDLDDVLTRLEPVARSLSCAD
MUL_1380|M.ulcerans_Agy99 VQENKWRAARYGLDAVIILDAESNERLVTDDLDDVLTRLEPVARSLSCAD
MAV_4721|M.avium_104 HQENKWRAARYGLDAVIILDADSNERLVTEDLDDVLNRLEPVARKLQCAD
Mb0441|M.bovis_AF2122/97 VQENKWRAARYGLDAVIILDADSNERLVTDDLADVLTRLEPVAKSLNCAD
Rv0433|M.tuberculosis_H37Rv VQENKWRAARYGLDAVIILDADSNERLVTDDLADVLTRLEPVAKSLNCAD
MSMEG_0836|M.smegmatis_MC2_155 VQENKWRAARYGLDAVIILDADSNERLVTEDLDDLLNHLEPVAASLHCAD
TH_0948|M.thermoresistible__bu VQENKWRAARYGLDAEIILDADSNERLVTDDLDDILNRLEPVAVSLNCAD
Mflv_0171|M.gilvum_PYR-GCK VQENKWRAARYGLDAIIILDADSNERLVTEDLDDLLERLQPVARKLDCVD
Mvan_0736|M.vanbaalenii_PYR-1 VQENKWRAARYGLDAIIILDADSNERLVTEDLDDLLERLQPVAKRLSCVE
MAB_4183c|M.abscessus_ATCC_199 VQENKWRSARYGLDAVIILDEDSNERLVTEDLDDLLTRLQPIARLLECET
******:******* **** :*******:** *:* :*:*:* * *
MMAR_0748|M.marinum_M ELAAVAEIPRVGASYQRQRRVAEEHDGDLRAVVDALIAELDI--------
MUL_1380|M.ulcerans_Agy99 ELAAVAEIPRVGASYQRQRRVAEEHDGDLRAVVDALIAELDI--------
MAV_4721|M.avium_104 ELAAVADIPRHGASYQRQRRVAEEHDGDLRAVVDALVAELEI--------
Mb0441|M.bovis_AF2122/97 ELAAVSDIYRDGASYQRQLRVAQQHDGDLRAVVDALVAELVI--------
Rv0433|M.tuberculosis_H37Rv ELAAVSDIYRDGASYQRQLRVAQQHDGDLRAVVDALVAELVI--------
MSMEG_0836|M.smegmatis_MC2_155 ELAAVEDIYRLGGSYQRQRRVAEENDGDLREVVDALIGELEL--------
TH_0948|M.thermoresistible__bu ELAEVEHIYRTGASYQRQRRVAEEHDGDLRAVVDALIGELDLGEPEQQGA
Mflv_0171|M.gilvum_PYR-GCK ELARVPDIYHNGASYQRQRRVAEEHDGDLRAVVDALVSELVL--------
Mvan_0736|M.vanbaalenii_PYR-1 ELSRVPDIYHNGASYQRQRRVAEEHDGDLRAVVDALVSELVL--------
MAB_4183c|M.abscessus_ATCC_199 ELAAVEEIYRHGGSYQRQRRVAEEHDGDLGAVVDAVVNELEPVRYTDSQ-
**: * .* : *.***** ***:::**** ****:: **
MMAR_0748|M.marinum_M -
MUL_1380|M.ulcerans_Agy99 -
MAV_4721|M.avium_104 -
Mb0441|M.bovis_AF2122/97 -
Rv0433|M.tuberculosis_H37Rv -
MSMEG_0836|M.smegmatis_MC2_155 -
TH_0948|M.thermoresistible__bu S
Mflv_0171|M.gilvum_PYR-GCK -
Mvan_0736|M.vanbaalenii_PYR-1 -
MAB_4183c|M.abscessus_ATCC_199 -