For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CGIVGYVGQQPACAVVMAALRRMEYRGYDSSGVALVNGDGTLTVSRRAGRLANLEEAVAQLPPSALTGTT GLGHTRWATHGRPTDRNAHPHRDAAGKIAVVHNGIIENYAGLRHELEADGVEFASDTDTEVAVHLVAQAY RHGLTAGDFAASVLAVLRRLDGHFTLVFANADEPGTIVAARRSTPLVIGIGDGEMFVGSDVAAFIPHTRN AIELGQDQAVVLTADGYRITDFDGNDDLGPGAYREFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLL GHFVDGRIVLDEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASEFRYRDPVLDR STLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQIPRECDAVLYTRAGPEIGVASTKTFLAQIT ANYLVGLALAQARGTKYPDEVEREYHELEAMPDLVARVLATIKPVAALAQRFAQSPTVLFLGRHVGYPVA LEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVIMPSPKGSAVLHAKLLSNIREIQARGAITIV IAEEGDDTVRPYADHLIEIPSVSTLLQPLLSTIPLQVFAASVAQARGYDVDKPRNLAKSVTVE*
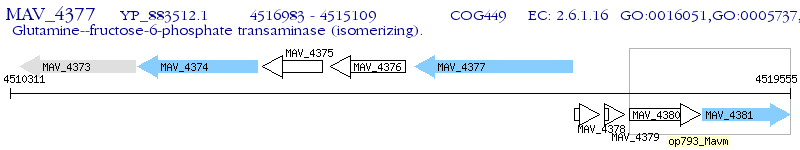
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4377 | glmS | - | 100% (624) | D-fructose-6-phosphate amidotransferase |
| M. avium 104 | MAV_0747 | purF | 7e-10 | 26.12% (245) | amidophosphoribosyltransferase |
| M. avium 104 | MAV_1188 | - | 2e-06 | 27.81% (169) | glutamine amidotransferase, class-II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3466c | glmS | 0.0 | 90.08% (625) | glucosamine--fructose-6-phosphate aminotransferase |
| M. gilvum PYR-GCK | Mflv_4939 | - | 0.0 | 86.22% (624) | D-fructose-6-phosphate amidotransferase |
| M. tuberculosis H37Rv | Rv3436c | glmS | 0.0 | 90.08% (625) | glucosamine--fructose-6-phosphate aminotransferase |
| M. leprae Br4923 | MLBr_00371 | glmS | 0.0 | 87.56% (627) | glucosamine--fructose-6-phosphate aminotransferase |
| M. abscessus ATCC 19977 | MAB_3743c | - | 0.0 | 82.21% (624) | glucosamine--fructose-6-phosphate aminotransferase |
| M. marinum M | MMAR_1114 | glmS | 0.0 | 90.56% (625) | glucosamine-fructose-6-phosphate aminotransferase, GlmS |
| M. smegmatis MC2 155 | MSMEG_1568 | glmS | 0.0 | 85.67% (628) | D-fructose-6-phosphate amidotransferase |
| M. thermoresistible (build 8) | TH_1701 | glmS | 0.0 | 86.06% (624) | PROBABLE GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_0872 | glmS | 0.0 | 90.56% (625) | glucosamine--fructose-6-phosphate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_1480 | - | 0.0 | 86.22% (624) | D-fructose-6-phosphate amidotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4939|M.gilvum_PYR-GCK MAMCGIVGYVGQRPACDIVVDALRRMEYRGYDSSGVALVDGH---GGLTV
Mvan_1480|M.vanbaalenii_PYR-1 --MCGIVGYVGQRPACDIVVDALRRMEYRGYDSAGVALLDGR---GGLTV
Mb3466c|M.bovis_AF2122/97 --MCGIVGYVGRRPAYVVVMDALRRMEYRGYDSSGIALVDG----GTLTV
Rv3436c|M.tuberculosis_H37Rv --MCGIVGYVGRRPAYVVVMDALRRMEYRGYDSSGIALVDG----GTLTV
MMAR_1114|M.marinum_M --MCGIVGYVGQRPACEVVLDALRRMEYRGYDSSGIALVDGS---GKLTV
MUL_0872|M.ulcerans_Agy99 --MCGIVGYVGQRPACEVVLDALRRMEYRGYDSSGIALVDGS---GKLTV
MLBr_00371|M.leprae_Br4923 --MCGLVGYVGQRPACGVVMDALRRMEYRGYDSSGIALINGSAKSGNLTV
MAV_4377|M.avium_104 --MCGIVGYVGQQPACAVVMAALRRMEYRGYDSSGVALVNGD---GTLTV
MSMEG_1568|M.smegmatis_MC2_155 --MCGIVGYVGHRPARDIVVDALRRMEYRGYDSAGIALIDGN---GGLTV
TH_1701|M.thermoresistible__bu --MCGIVGYVGARNALDIVVDALRRMEYRGYDSAGVALLDGA---GGLTV
MAB_3743c|M.abscessus_ATCC_199 --MCGIVGYVGHRDALSVVLEALRRLEYRGYDSAGVALADGH---GGLLV
***:***** : * :*: ****:*******:*:** :* * * *
Mflv_4939|M.gilvum_PYR-GCK RRKAGRLANLEAALAEAGA---ESPAGATGLGHTRWATHGRPTDRNAHPH
Mvan_1480|M.vanbaalenii_PYR-1 RRKAGRLANLEAALAEGGT---DNLVGATGLGHTRWATHGRPTDRNAHPH
Mb3466c|M.bovis_AF2122/97 RRRAGRLANLEEAVAEMPS---TALSGTTGLGHTRWATHGRPTDRNAHPH
Rv3436c|M.tuberculosis_H37Rv RRRAGRLANLEEAVAEMPS---TALSGTTGLGHTRWATHGRPTDRNAHPH
MMAR_1114|M.marinum_M RRRAGRLANLETAVAEMSA---ESLTGTTGLGHTRWATHGRPTDRNAHPH
MUL_0872|M.ulcerans_Agy99 RRRAGRLANLETAVAEMPA---ESLTGTTGLGHTRWATHGRPTDRNAHPH
MLBr_00371|M.leprae_Br4923 RRRAGRLSNLESVLAEMVP---ASLAGNVGLGHIRWATHGRPTDRNAHPH
MAV_4377|M.avium_104 SRRAGRLANLEEAVAQLPP---SALTGTTGLGHTRWATHGRPTDRNAHPH
MSMEG_1568|M.smegmatis_MC2_155 RRRAGRLANLEATLAETDSNDGDGLGGSTGLGHTRWATHGRPTDRNAHPH
TH_1701|M.thermoresistible__bu RRRAGRLANLEAELAATDP---GLLAGTAGLGHTRWATHGRPTDRNAHPH
MAB_3743c|M.abscessus_ATCC_199 QRKAGRLANLESAIAESGE----SFAATTGMGHTRWATHGAPTDRNAHPH
*:****:*** :* . .*:** ****** *********
Mflv_4939|M.gilvum_PYR-GCK RDSSGKLAVVHNGIIENFAALRAELEAVGVEFASDTDTEVAVHLVAREYT
Mvan_1480|M.vanbaalenii_PYR-1 RDASGKVAVVHNGIIENFAVLRAELEAAGVEFASDTDTEVTVHLVAREYV
Mb3466c|M.bovis_AF2122/97 RDAAGKIAVVHNGIIENFAVLRRELETAGVEFASDTDTEVAAHLVARAYR
Rv3436c|M.tuberculosis_H37Rv RDAAGKIAVVHNGIIENFAVLRRELETAGVEFASDTDTEVAAHLVARAYR
MMAR_1114|M.marinum_M RDAAGKIAVVHNGIIENFAVLRQELEAAGVEFASDTDTEVAVHLVAQAFH
MUL_0872|M.ulcerans_Agy99 RDAAGKIAVVHNGIIENFAVLRQELEAAGVEFASDTDTEVAVHLVAQAFH
MLBr_00371|M.leprae_Br4923 RDATGKIAVVHNGIIENFPSLRHELEIAGVEFVSDTDTEVAVHLVAQAYC
MAV_4377|M.avium_104 RDAAGKIAVVHNGIIENYAGLRHELEADGVEFASDTDTEVAVHLVAQAYR
MSMEG_1568|M.smegmatis_MC2_155 RDAAGKIAVVHNGIIENFAPLRAELEAAGVEFASDTDTEVAVHLVARQYT
TH_1701|M.thermoresistible__bu RDASGKFAVAHNGIIENNAVLRAELEAAGVEFASDTDTEVAVHLLARAYQ
MAB_3743c|M.abscessus_ATCC_199 QDATGKVAVVHNGIIENFAVLRAELESAGVEFASDTDSEVAVHLVSRQFE
:*::**.**.******* . ** *** ****.****:**:.**::: :
Mflv_4939|M.gilvum_PYR-GCK HGDTAGDFPASVFAVLPRLEGHFTLVFAHADDPGTIVAARRSTPLVLGVG
Mvan_1480|M.vanbaalenii_PYR-1 HGETAGDFPASVLAVLRRLEGHFTLVFAHADDPGTIVAARRSTPLVLGVG
Mb3466c|M.bovis_AF2122/97 HGETADDFVGSVLAVLRRLEGHFTLVFANADDPGTLVAARRSTPLVLGIG
Rv3436c|M.tuberculosis_H37Rv HGETADDFVGSVLAVLRRLEGHFTLVFANADDPGTLVAARRSTPLVLGIG
MMAR_1114|M.marinum_M HGETAGDFPASVLSVLRRLEGHFTLVFANADDPGTIVAARRSTPLVLGIG
MUL_0872|M.ulcerans_Agy99 HGETAGDFPASVLSVLRRLEGHFTLVFANADDPGTIVAARRSTPLVLGIG
MLBr_00371|M.leprae_Br4923 AGETAGDFVGSVLAVLRRLQGHFTLVFANADEPGTIVAARRSTPLVLGIG
MAV_4377|M.avium_104 HGLTAGDFAASVLAVLRRLDGHFTLVFANADEPGTIVAARRSTPLVIGIG
MSMEG_1568|M.smegmatis_MC2_155 QGDTAGDFPASVLAVLQRLEGHFTLVFASADDPGTIVAARRSTPLVLGIG
TH_1701|M.thermoresistible__bu HGETAGDFVASVLSVLRRLDGHFTLVFANADEPGTIIAARRSTPLVVGVG
MAB_3743c|M.abscessus_ATCC_199 SGDTAGDFVASVQSVVRRLEGHFTLVFSHADDPGTIVAARRSTPLVVGIG
* **.** .** :*: **:*******: **:***::*********:*:*
Mflv_4939|M.gilvum_PYR-GCK DGEMFVGSDVAAFIEHTREAVELGQDQAVVVTADGYRVTDFHGNDDAAS-
Mvan_1480|M.vanbaalenii_PYR-1 DGEMFVGSDVAAFIEHTRDAVELGQDQAVVVTADGYRVTDFHGNPDDAR-
Mb3466c|M.bovis_AF2122/97 DNEMFVGSDVAAFIEHTREAVELGQDQAVVITADGYRISDFDGNDGLQAG
Rv3436c|M.tuberculosis_H37Rv DNEMFVGSDVAAFIEHTREAVELGQDQAVVITADGYRISDFDGNDGLQAG
MMAR_1114|M.marinum_M DGEMFVGSDVAAFITHTRHAVELGQDQAVVITADGYRISDFDGNDDLLAG
MUL_0872|M.ulcerans_Agy99 DGEMFVGSDVAAFITHTRHAVDLGQDQAVVITADGYRISDFDGNDDLLAG
MLBr_00371|M.leprae_Br4923 DGEMFVGSDVAAFIEHTRQAVELGQDQAVVITADGYRISDFDGNDDAVN-
MAV_4377|M.avium_104 DGEMFVGSDVAAFIPHTRNAIELGQDQAVVLTADGYRITDFDGNDDLGPG
MSMEG_1568|M.smegmatis_MC2_155 DGEMFVGSDVAAFIEHTRDAVELGQDQAVVLTADGYRITDFAGNDHLEAG
TH_1701|M.thermoresistible__bu DGEMFVGSDVAAFIEHTRDAVELGQDQAVVITADSYRVLDFLGADVTAQ-
MAB_3743c|M.abscessus_ATCC_199 DGEMFLGSDVAAFIEYTRDAVELGQDQVVVITADGYRITDFEGNDDTGN-
*.***:******** :**.*::*****.**:***.**: ** *
Mflv_4939|M.gilvum_PYR-GCK --ARPFHIDWDLSAAEKGGYEYFMLKEIAEQPAAVSDTLLGHFVDNRIVL
Mvan_1480|M.vanbaalenii_PYR-1 --TRRFHIDWDTSAAEKGGYEYFMLKEIAEQPAAVSDTLLGHFADNRIVL
Mb3466c|M.bovis_AF2122/97 RDFRPFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVGGRIVL
Rv3436c|M.tuberculosis_H37Rv RDFRPFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVGGRIVL
MMAR_1114|M.marinum_M TDYREFHIDWDLAAAEKGGYEYFMLKEIAEQPTAVAETLLGHFVGNRIVL
MUL_0872|M.ulcerans_Agy99 TDYREFHIDWDLAAAEKGGYEYFMLKEIAEQPTAVAETLLGHFVGNRIVL
MLBr_00371|M.leprae_Br4923 --ARTFHIDWDLAAAEKGGYEYFMLKEIAEQPDAVVDTLLGHFTGGRIVL
MAV_4377|M.avium_104 -AYREFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVDGRIVL
MSMEG_1568|M.smegmatis_MC2_155 RDFREFHIDWDLNAAEKGGYDYFMLKEIAEQPSAVADTLLGHFDKNRIVL
TH_1701|M.thermoresistible__bu --ARHFHIDWDLSAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVDGRIVL
MAB_3743c|M.abscessus_ATCC_199 --ARVFTIDWDLSAAEKGGYEYFMLKEIAEQPAAVSDTLLGHFDLDRIVL
* * **** *******:*********** ** :****** .****
Mflv_4939|M.gilvum_PYR-GCK DEQRLSDQELRDIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Mvan_1480|M.vanbaalenii_PYR-1 DEQRLSDQELRDIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Mb3466c|M.bovis_AF2122/97 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Rv3436c|M.tuberculosis_H37Rv DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MMAR_1114|M.marinum_M DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MUL_0872|M.ulcerans_Agy99 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MLBr_00371|M.leprae_Br4923 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYTIEHWTRLPVEVELASE
MAV_4377|M.avium_104 DEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MSMEG_1568|M.smegmatis_MC2_155 DEQRLSDQELREIDKVFIVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
TH_1701|M.thermoresistible__bu DEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MAB_3743c|M.abscessus_ATCC_199 DEQRLSDQELRDIDKVFVVACGTAFHSGLLAKYAIEHWTRLPVEIELASE
***********::****:******:********:**********:*****
Mflv_4939|M.gilvum_PYR-GCK FRYRDPVLDSHTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
Mvan_1480|M.vanbaalenii_PYR-1 FRYRDPVLDSHTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
Mb3466c|M.bovis_AF2122/97 FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
Rv3436c|M.tuberculosis_H37Rv FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
MMAR_1114|M.marinum_M FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
MUL_0872|M.ulcerans_Agy99 FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
MLBr_00371|M.leprae_Br4923 FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
MAV_4377|M.avium_104 FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGSQ
MSMEG_1568|M.smegmatis_MC2_155 FRYRDPVLDRSTLVIAISQSGETADTLEAVRHAKTQKAKVLAICNTNGSQ
TH_1701|M.thermoresistible__bu FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKAQKAKVLAICNTNGSQ
MAB_3743c|M.abscessus_ATCC_199 FRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKEQKAKVLAVCNTNGSQ
********* ***:******************* *******:*******
Mflv_4939|M.gilvum_PYR-GCK IPRECDAVLYTRAGPEIGVASTKTFLAQVAANYLVGLALAQARGTKYPDE
Mvan_1480|M.vanbaalenii_PYR-1 IPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLVGLALAQARGTKYPDE
Mb3466c|M.bovis_AF2122/97 IPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLLGLALAQARGTKYPDE
Rv3436c|M.tuberculosis_H37Rv IPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLLGLALAQARGTKYPDE
MMAR_1114|M.marinum_M IPRECDAVLYTRAGPEIGVASTKTFLAQVTANYLLGLALAQARGTKYPDE
MUL_0872|M.ulcerans_Agy99 IPRECDAVLYTRAGPEIGVASTKTFLAQVTANYLLGLALAQARGTKYPDE
MLBr_00371|M.leprae_Br4923 IPRECDAVLYTRAGPEIGVASTKTFLAQVAANYLLGLALAQARGTKYPDE
MAV_4377|M.avium_104 IPRECDAVLYTRAGPEIGVASTKTFLAQITANYLVGLALAQARGTKYPDE
MSMEG_1568|M.smegmatis_MC2_155 IPREADAVLYTRAGPEIGVAATKTFLAQIAANYLVGLALAQARGTKYPDE
TH_1701|M.thermoresistible__bu IPRESDAVLYTRAGPEIGVAATKTFLAQIVANYLVGLALAQARGTKYPDE
MAB_3743c|M.abscessus_ATCC_199 IPRECDAVLYTRAGPEIGVAATKTFLAQVTANYIVGLALAQARGTKYPDE
****.***************:*******:.***::***************
Mflv_4939|M.gilvum_PYR-GCK VAREYHDLEAMPDLISRVLASSEPVVALAKQFATSQTVLFLGRHVGYPVA
Mvan_1480|M.vanbaalenii_PYR-1 VAREYHDLEAMPDLIARVLDTSEPVLQLAKQFAASQTVLFLGRHVGYPVA
Mb3466c|M.bovis_AF2122/97 VEREYHELEAMPDLVARVIAATGPVAELAHRFAQSSTVLFLGRHVGYPVA
Rv3436c|M.tuberculosis_H37Rv VEREYHELEAMPDLVARVIAATGPVAELAHRFAQSSTVLFLGRHVGYPVA
MMAR_1114|M.marinum_M VEREYRELEAMPDLVARVISAIKPVADLAYRFAQSSTVLFLGRHVGYPVA
MUL_0872|M.ulcerans_Agy99 VEREYRELEAMPDLVARVISAIKPVAHLAYRFAQSSTVLFLGRHVGYPVA
MLBr_00371|M.leprae_Br4923 VQREYRELEAMPDLVARVIAGMGPVADLAYRFAQSTTVLFLGRHVGYPVA
MAV_4377|M.avium_104 VEREYHELEAMPDLVARVLATIKPVAALAQRFAQSPTVLFLGRHVGYPVA
MSMEG_1568|M.smegmatis_MC2_155 VAREYRELEAMPDLIKRVLAGMDSVAALAERFAPSSTVLFLGRHVGYPVA
TH_1701|M.thermoresistible__bu VEREYRELEAMPALVQRVVSTVEPVSALARQFAGAPAVLFLGRHVGYPVA
MAB_3743c|M.abscessus_ATCC_199 VAREYHELEAMPELIERVISQMDPVADLARQYAQSSSILFLGRHVGYPVA
* ***::***** *: **: .* ** ::* : ::************
Mflv_4939|M.gilvum_PYR-GCK LEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVVMPSPKNSATL
Mvan_1480|M.vanbaalenii_PYR-1 LEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVVMPSPKNSATL
Mb3466c|M.bovis_AF2122/97 LEGALKLKELAYMHAEGFAAGELKHGPIALIEDGLPVIVVMPSPKGSATL
Rv3436c|M.tuberculosis_H37Rv LEGALKLKELAYMHAEGFAAGELKHGPIALIEDGLPVIVVMPSPKGSATL
MMAR_1114|M.marinum_M LEGALKLKELAYMHAEGFAAGELKHGPIALVEDELPVIVIMPSPKGSATL
MUL_0872|M.ulcerans_Agy99 LEGALKLKELAYMHAEGFAAGELKHGPIALVEDELPVIVIMPSPKGSATL
MLBr_00371|M.leprae_Br4923 LEGALKLKELAYMHAEGFAAGELKHGPIALIEENLPVIVVMPSPKGSAML
MAV_4377|M.avium_104 LEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVIMPSPKGSAVL
MSMEG_1568|M.smegmatis_MC2_155 LEGALKLKELAYMHAEGFAAGELKHGPIALIDENLPVIVVMPSPKNAAML
TH_1701|M.thermoresistible__bu LEGALKLKELAYMHAEGFAAGELKHGPIALIEEGLPVIVVMPSPKHALTL
MAB_3743c|M.abscessus_ATCC_199 LEGALKLKELAYMHAEGFAAGELKHGPIALIEEGLPVIVVMPSPKGMGLL
******************************::: *****:***** *
Mflv_4939|M.gilvum_PYR-GCK HSKLLSNIREIQARGAVTIVIAEEGDETVRPYADHLIEIPAVSTLFQPLL
Mvan_1480|M.vanbaalenii_PYR-1 HSKLLSNIREIQARGAVTIVIAEEGDDTVRPYADHLIEIPAVSTLFQPLL
Mb3466c|M.bovis_AF2122/97 HAKLLSNIREIQTRGAVTIVIAEEGDETVRPYADHLIEIPAVSTLLQPLL
Rv3436c|M.tuberculosis_H37Rv HAKLLSNIREIQTRGAVTIVIAEEGDETVRPYADHLIEIPAVSTLLQPLL
MMAR_1114|M.marinum_M HAKLLSNIREIQTRGAVTIVIAEEGDDTVRPFADHLIEIPAVSTLLQPLL
MUL_0872|M.ulcerans_Agy99 HAKLLSNIREIQTRGAVTIVIAEEGDDTVRPFADHLIEIPAVSTLLQPLL
MLBr_00371|M.leprae_Br4923 HAKLLSNIREIQTRGAVTIVIAEEGDDTVRLYADHLIELPAVSTLLQPLL
MAV_4377|M.avium_104 HAKLLSNIREIQARGAITIVIAEEGDDTVRPYADHLIEIPSVSTLLQPLL
MSMEG_1568|M.smegmatis_MC2_155 HAKLLSNIREIQARGAVTVVIAEEDDDTVRPYADHLIEIPSVSTLFQPLL
TH_1701|M.thermoresistible__bu HGKLLSNIREIQARGAVTIVIAEEGDDTVRPYADHLIEIPVVSTLFQPLL
MAB_3743c|M.abscessus_ATCC_199 HSKLLSNIREIQARGARTIVIAEEGDETIRPYADHLIEIPAVSTLYQPLL
*.**********:*** *:*****.*:*:* :******:* **** ****
Mflv_4939|M.gilvum_PYR-GCK STIPMQLFSAGVAQARGYDVDKPRNLAKSVTVE
Mvan_1480|M.vanbaalenii_PYR-1 STIPMQLFSAGVAQARGYDVDKPRNLAKSVTVE
Mb3466c|M.bovis_AF2122/97 STIPLQVFAASVARARGYDVDKPRNLAKSVTVE
Rv3436c|M.tuberculosis_H37Rv STIPLQVFAASVARARGYDVDKPRNLAKSVTVE
MMAR_1114|M.marinum_M STIPLQVFAAAVAQARGYDVDKPRNLAKSVTVE
MUL_0872|M.ulcerans_Agy99 STIPLQVFAAAVAQARGYDVDKPRNLAKSVTVE
MLBr_00371|M.leprae_Br4923 STIPLQVFAASVAQARGYDVDKPRNLAKSVTVE
MAV_4377|M.avium_104 STIPLQVFAASVAQARGYDVDKPRNLAKSVTVE
MSMEG_1568|M.smegmatis_MC2_155 STIPLQVFAAGVARARGYDVDKPRNLAKSVTVE
TH_1701|M.thermoresistible__bu STVPLQVFAASVAQARGYDVDKPRNLAKSVTVE
MAB_3743c|M.abscessus_ATCC_199 STIPMQVFAAAVAQARGYDVDKPRNLAKSVTVE
**:*:*:*:*.**:*******************