For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHKGATFAVVAVTAAIAPLAACANQQSSQPNTAPLTSSVPGSERLTTQLKTAEGIPVANASFEFANGYAT VTVEAGPNQVLSPGFHGLQIHAVGKCEANSTAPTGGSTGDFESAGAVYQAPDHTGYPASGDLTALQVRSD GSAKLVTTSNAFTAADLRTSSGSALILHQNANNLANTPAADSGKRLACGVIAASSATSTTTTPTTSVTTS TTTVAVPPPSTSTSTSTVTVTGTPTATSTPTTTVTTPPSLPPGR
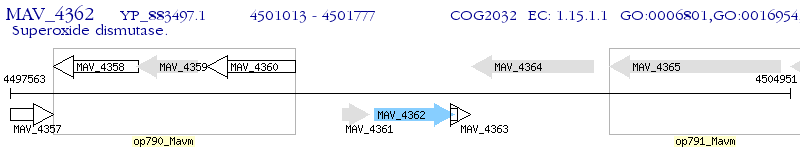
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4362 | sodC | - | 100% (254) | superoxide dismutase, Cu-Zn |
| M. avium 104 | MAV_4722 | sodC | 7e-49 | 47.49% (219) | superoxide dismutase, Cu-Zn |
| M. avium 104 | MAV_2043 | - | 2e-42 | 47.98% (198) | [Cu-Zn] superoxide dismutase [Mycobacterium avium 104] |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0440 | sodC | 8e-46 | 46.98% (215) | periplasmic superoxide dismutase |
| M. gilvum PYR-GCK | Mflv_1989 | - | 9e-53 | 45.35% (258) | superoxide dismutase, copper/zinc binding |
| M. tuberculosis H37Rv | Rv0432 | sodC | 8e-46 | 46.98% (215) | periplasmic superoxide dismutase |
| M. leprae Br4923 | MLBr_01925 | sodC | 7e-41 | 42.86% (217) | superoxide dismutase precursor (Cu-Zn) |
| M. abscessus ATCC 19977 | MAB_4184c | - | 1e-43 | 41.45% (234) | superoxide dismutase |
| M. marinum M | MMAR_0747 | sodC | 2e-41 | 44.05% (227) | periplasmic superoxide dismutase |
| M. smegmatis MC2 155 | MSMEG_0835 | sodC | 1e-47 | 44.49% (236) | copper/zinc superoxide dismutase |
| M. thermoresistible (build 8) | TH_0050 | sodC | 2e-58 | 45.04% (282) | PROBABLE PERIPLASMIC SUPEROXIDE DISMUTASE |
| M. ulcerans Agy99 | MUL_1379 | sodC | 2e-41 | 43.17% (227) | periplasmic superoxide dismutase |
| M. vanbaalenii PYR-1 | Mvan_0735 | - | 2e-45 | 53.53% (170) | superoxide dismutase, copper/zinc binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0747|M.marinum_M MPKPAGHRNVAAVTMSALS---ATACVALLSGCSS-PQHASTVPGTTPSV
MUL_1379|M.ulcerans_Agy99 MPKPAGHRNVAAVTMSALS---ATACVALLSGCSS-PQHASTVPGTTPSV
MLBr_01925|M.leprae_Br4923 MSKLAGHRNVAAVTRSALSLSFVAACVALLSACIQ-NQPPATLPGTTPTV
Mb0440|M.bovis_AF2122/97 MPKPADHRNHAAVSTSVLSALFLGAGAALLSACSS-PQHASTVPGTTPSI
Rv0432|M.tuberculosis_H37Rv MPKPADHRNHAAVSTSVLSALFLGAGAALLSACSS-PQHASTVPGTTPSI
MSMEG_0835|M.smegmatis_MC2_155 -----------MLKPVSVAV-LFATPVLALSACSPPGETASSEPGTTPAI
Mvan_0735|M.vanbaalenii_PYR-1 -----------MLRSVAAAT-LFAVPALALSACSPPGEVPSDSPGTTPPV
MAB_4184c|M.abscessus_ATCC_199 --------MKPMVKPAVLASGTLAAVALSLSGCSP-DQQASTTPGTTPPV
TH_0050|M.thermoresistible__bu -----MTNPKQVPRPAIAAALAAPVALALLTGCQQDGQSAEDTTSPT---
MAV_4362|M.avium_104 -----------MHKGATFAVVAVTAAIAPLAACAN--Q--QSSQPNTAPL
Mflv_1989|M.gilvum_PYR-GCK -----------MNKHVTAAIALLATPAAILSGCSN--QSGEDESSTTTPS
: . *:.* : *
MMAR_0747|M.marinum_M WTGSPAPAGTNEHREGAS-------ESEGLATQLRAPDGTEVATAKFDFA
MUL_1379|M.ulcerans_Agy99 WTGSPASAGTNEHREGAS-------ESEGLATQLRAPDGAEVATAKFDFA
MLBr_01925|M.leprae_Br4923 WTGSPAPSGMLGAEAESM-------GPPNIITRLNAPDGTQVATAKFEFN
Mb0440|M.bovis_AF2122/97 WTGSPAPSGLSGHDEESP-------GAQSLTSTLTAPDGTKVATAKFEFA
Rv0432|M.tuberculosis_H37Rv WTGSPAPSGLSGHDEESP-------GAQSLTSTLTAPDGTKVATAKFEFA
MSMEG_0835|M.smegmatis_MC2_155 WTGSPSPAAPSGEDHGGGHGAGAAGAGETLTAELKTADGTSVATADFQFA
Mvan_0735|M.vanbaalenii_PYR-1 WTGSPSPTAGPDE-QGSGHGSGTQAGGETLTADLKLADGTTVATADITFS
MAB_4184c|M.abscessus_ATCC_199 WTGSTAPAT-AGEHGGHGGGEQPVPAGEKLNATLKLADGTPVATAAFVFQ
TH_0050|M.thermoresistible__bu -TTSPAPRG------------------ETISTELKTPDGRTVADADIEFT
MAV_4362|M.avium_104 TSS-VPGSE-------------------RLTTQLKTAEGIPVANASFEFA
Mflv_1989|M.gilvum_PYR-GCK SSANAADAQ-------------------QLKTQLKTVDDKAVAEATIDFT
: . : : * :. ** * : *
MMAR_0747|M.marinum_M DGFATVTIVTTGTGYLAPGFHGVHIHKVGKCEANSAGPTGGPVGDFLSAG
MUL_1379|M.ulcerans_Agy99 DGFATVTIVTTGTGYLAPGFHGVHIHKVGKCEANSAGPTGGPVGDFLSAG
MLBr_01925|M.leprae_Br4923 NGFATITIATTGVGHLAPGFHGVHIHKVGKCEPSSAGPTGGAPGDFLSAG
Mb0440|M.bovis_AF2122/97 NGYATVTIATTGVGKLTPGFHGLHIHQVGKCEPNSVAPTGGAPGNFLSAG
Rv0432|M.tuberculosis_H37Rv NGYATVTIATTGVGKLTPGFHGLHIHQVGKCEPNSVAPTGGAPGNFLSAG
MSMEG_0835|M.smegmatis_MC2_155 DGFATVTIETTTPGRLTPGFHGVHIHSVGKCEANSVAPTGGAPGDFNSAG
Mvan_0735|M.vanbaalenii_PYR-1 DGYATVTVEASGAGELTPGFHGIHIHSVGKCEADSVAPTGGAPADFNSAG
MAB_4184c|M.abscessus_ATCC_199 DGYARITVKTTQNGKLTPGFHGLHIHSFKKCEANSVAPAGGAPGDFLSAG
TH_0050|M.thermoresistible__bu DGYATITVETTESGILSPGFHGLHIHEIGKCEANSVAPGGGEPGDFLSAG
MAV_4362|M.avium_104 NGYATVTVEAGPNQVLSPGFHGLQIHAVGKCEANSTAPTGGSTGDFESAG
Mflv_1989|M.gilvum_PYR-GCK GGFATVTVETVDGATLSPGFHGLHIHAFGRCEPNSPAPDGGPVGDFNSAG
.*:* :*: : *:*****::** . :**..* .* ** .:* ***
MMAR_0747|M.marinum_M GHLQVSGRPVPGQSSGDLTSLQVRSDGSGTLVTTTDAFTMDDLTSGQKTA
MUL_1379|M.ulcerans_Agy99 GHLQVSGRPVPGQSSGDLTSLQVRSDGSGTLVTTTDAFTMDDLTSGQKTA
MLBr_01925|M.leprae_Br4923 GHFQVPGHTVEP-ASGNLTSLQVRKDGIGTLVTTTDAFTMNDLLAGQKTA
Mb0440|M.bovis_AF2122/97 GHYHVPGHTGTP-ASGDLASLQVRGDGSAMLVTTTDAFTMDDLLSGAKTA
Rv0432|M.tuberculosis_H37Rv GHYHVPGHTGTP-ASGDLASLQVRGDGSAMLVTTTDAFTMDDLLSGAKTA
MSMEG_0835|M.smegmatis_MC2_155 GHFQVSGHSGHP-ASGDLSSLQVRADGSGKLVTTTDAFTAEDLLDGAKTA
Mvan_0735|M.vanbaalenii_PYR-1 GHFQVSGHSGHP-ASGDLSSLQVREDGSAKLVTTTDAFTKEDLLDGAGTA
MAB_4184c|M.abscessus_ATCC_199 GHFNVPGYTGHP-SSGDLVSLNILKDGSGELVTTADSFTKEDLLAGNGTA
TH_0050|M.thermoresistible__bu GHLHMRGDTPHP-GTGDLTPLEVRSDGSARLVATTDSLTIEDLRGSQGSA
MAV_4362|M.avium_104 AVYQAPDHTGYP-ASGDLTALQVRSDGSAKLVTTSNAFTAADLRTSSGSA
Mflv_1989|M.gilvum_PYR-GCK DHFQGEGMTGMP-AAGDLPPLLVRSDGNGKLVATTDTFTEEQLTGPDGSS
: . . .:*:* .* : ** . **:*::::* :* ::
MMAR_0747|M.marinum_M IIIHAGADNFANIPAD----RYNQTNGTAGPDQTTLATGDAGKRVACGVI
MUL_1379|M.ulcerans_Agy99 IIIHAGADNFANIPAD----RYNQTNGTAGPDQTTLATGDAGKRVACGVI
MLBr_01925|M.leprae_Br4923 IIIHAGADNFGNIPPE----RYSQVNGTPGPDATTISTGDAGKRVACGVI
Mb0440|M.bovis_AF2122/97 IIIHAGADNFANIPPE----RYVQVNGTPGPDETTLTTGDAGKRVACGVI
Rv0432|M.tuberculosis_H37Rv IIIHAGADNFANIPPE----RYVQVNGTPGPDETTLTTGDAGKRVACGVI
MSMEG_0835|M.smegmatis_MC2_155 IIIHEKADNFANIPPE----RYQQVNGAPGPDQTTMATGDAGSRVACGVI
Mvan_0735|M.vanbaalenii_PYR-1 LIIHEKADNFANIPPE----RYQQVNGAPPPDQTTLATGDAGARVACGVI
MAB_4184c|M.abscessus_ATCC_199 IIIHEKSDNFANIPAE----RYTQANGASGPDDTTKSTGDAGKRVACGLI
TH_0050|M.thermoresistible__bu LMLHERPDNFANIPP-----RYT-VNGAPGPDAETLATGDAGSRIACGAL
MAV_4362|M.avium_104 LILHQNANNLANTPAADSGKRLACGVIA-ASSATSTTTTPTTSVTTSTTT
Mflv_1989|M.gilvum_PYR-GCK ILLHEGADMPGAEQGADT--RIACGVISPASTETSVSTSTSTSTVTTTVA
:::* .: . * : . : :* : :
MMAR_0747|M.marinum_M GSG-----------------------------------------------
MUL_1379|M.ulcerans_Agy99 GSG-----------------------------------------------
MLBr_01925|M.leprae_Br4923 GAD-----------------------------------------------
Mb0440|M.bovis_AF2122/97 GSG-----------------------------------------------
Rv0432|M.tuberculosis_H37Rv GSG-----------------------------------------------
MSMEG_0835|M.smegmatis_MC2_155 SAG-----------------------------------------------
Mvan_0735|M.vanbaalenii_PYR-1 TGG-----------------------------------------------
MAB_4184c|M.abscessus_ATCC_199 DAG-----------------------------------------------
TH_0050|M.thermoresistible__bu AAASAQATPTTETTTITETAPGGPPAGTAPPATEPPGTTGPPATTESPQT
MAV_4362|M.avium_104 VAVPP------------------------------------PSTSTSTST
Mflv_1989|M.gilvum_PYR-GCK PAVPPNT------------------------ASETTPTTPPPTTDSPTPT
.
MMAR_0747|M.marinum_M ----------------------------
MUL_1379|M.ulcerans_Agy99 ----------------------------
MLBr_01925|M.leprae_Br4923 ----------------------------
Mb0440|M.bovis_AF2122/97 ----------------------------
Rv0432|M.tuberculosis_H37Rv ----------------------------
MSMEG_0835|M.smegmatis_MC2_155 ----------------------------
Mvan_0735|M.vanbaalenii_PYR-1 ----------------------------
MAB_4184c|M.abscessus_ATCC_199 ----------------------------
TH_0050|M.thermoresistible__bu TTTTAETTVTTPPTTTVTTTVPEGEGPG
MAV_4362|M.avium_104 VTVTGTPTATSTPTTTVTTPPSLPPGR-
Mflv_1989|M.gilvum_PYR-GCK TTVTTETTAPTETVKTTTTAPVPAPNG-