For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDTFHDPVDHLRTTRPLAGESLIDVLHWPGYLLVVAGVVGACGSLAAFATGHHHEGMTAGVTAVIAAVLG LVWLAVEHRRIRRIADRWYAEHPEVHRQWPAS*
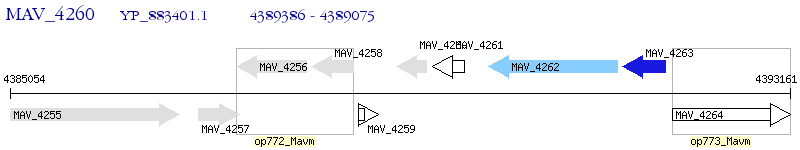
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4260 | - | - | 100% (103) | UsfY protein |
| M. avium 104 | MAV_0438 | - | 8e-10 | 34.83% (89) | hypothetical protein MAV_0438 |
| M. avium 104 | MAV_2939 | - | 3e-07 | 33.71% (89) | hypothetical protein MAV_2939 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3316c | usfY | 2e-45 | 78.79% (99) | hypothetical protein Mb3316c |
| M. gilvum PYR-GCK | Mflv_4793 | - | 2e-23 | 50.52% (97) | putative protein UsfY |
| M. tuberculosis H37Rv | Rv3288c | usfY | 2e-45 | 78.79% (99) | hypothetical protein Rv3288c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2519c | - | 1e-17 | 43.30% (97) | hypothetical protein MAB_2519c |
| M. marinum M | MMAR_1245 | usfY | 2e-46 | 79.61% (103) | hypothetical protein MMAR_1245 |
| M. smegmatis MC2 155 | MSMEG_1791 | - | 2e-25 | 54.64% (97) | UsfY protein |
| M. thermoresistible (build 8) | TH_1001 | - | 7e-18 | 45.16% (93) | UsfY protein |
| M. ulcerans Agy99 | MUL_2642 | usfY | 8e-46 | 78.64% (103) | hypothetical protein MUL_2642 |
| M. vanbaalenii PYR-1 | Mvan_1655 | - | 3e-21 | 46.94% (98) | putative protein UsfY |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4793|M.gilvum_PYR-GCK -----------------------------------MRGPSDPVDHVRTTR
Mvan_1655|M.vanbaalenii_PYR-1 -----------------------------------MKGPNDPVDHVRTTR
MSMEG_1791|M.smegmatis_MC2_155 -----------------------------------MKGPKDPVDHARTTR
TH_1001|M.thermoresistible__bu ------------------------------------------VDHARTTR
MMAR_1245|M.marinum_M ----------------------------------MGDTYRDPVDHLRTTR
MUL_2642|M.ulcerans_Agy99 ----------------------------------MGDTYRDPVDHLRTTR
Mb3316c|M.bovis_AF2122/97 MGQIPPQPVRRVLPLMVVPGNGQKWRNRTETEEAMGDTYRDPVDHLRTTR
Rv3288c|M.tuberculosis_H37Rv MGQIPPQPVRRVLPLMVVPGNGQKWRNRTETEEAMGDTYRDPVDHLRTTR
MAV_4260|M.avium_104 ----------------------------------MGDTFHDPVDHLRTTR
MAB_2519c|M.abscessus_ATCC_199 ----------------------------------MHDDGIDPVDHARTFR
*** ** *
Mflv_4793|M.gilvum_PYR-GCK PHAGESMKDNKIMPGLVVIGLSLVSFVAMLSAFATSHHDVGVMLGAIAAA
Mvan_1655|M.vanbaalenii_PYR-1 PHAGESMKDNKIMPGLVVIGLALVSFVSALAAFATVNHDVGVMLGALGLA
MSMEG_1791|M.smegmatis_MC2_155 PHAGESMKDNMIMPALIVIGLALVTFVGSLAAFATKHHDVGLTLVSLAAA
TH_1001|M.thermoresistible__bu PHAGESLKNTASMPALIVLGLGLVAFVSCLATLAAGETMAGVVLACLAVL
MMAR_1245|M.marinum_M PLAGESLIDVLHWPGYLLVVAGVIGGVGSLAAFGTGHHSEGMASGVVALT
MUL_2642|M.ulcerans_Agy99 PLAGESLIDVLHWPGYLLVVAGVIGGVGSLAAFGTGHHSEGMASGVVALT
Mb3316c|M.bovis_AF2122/97 PLAGESLIDVVHWPGYLLIVAGVVGGVGALAAFGTGHHAEGMTFGVVAIV
Rv3288c|M.tuberculosis_H37Rv PLAGESLIDVVHWPGYLLIVAGVVGGVGALAAFGTGHHAEGMTFGVVAIV
MAV_4260|M.avium_104 PLAGESLIDVLHWPGYLLVVAGVVGACGSLAAFATGHHHEGMTAGVTAVI
MAB_2519c|M.abscessus_ATCC_199 PHAGESLKDRVAWPGLAFIALGVLSVVLGLAAAAYMFYGWTLVAGIAALV
* ****: : *. .: .:: *:: . : .
Mflv_4793|M.gilvum_PYR-GCK GFVLGGGWLFIEHQRVRRLEERWYAEHPGAWRQRP----NS
Mvan_1655|M.vanbaalenii_PYR-1 GFVVGGGWLMIEHHRVRHLEDVWYAQHPGVPRQRP----NS
MSMEG_1791|M.smegmatis_MC2_155 GFVVGALWLALEHLRVRRIEERWYAEHPGVMRQGP----SS
TH_1001|M.thermoresistible__bu GFAVAGVWLMVEHRRVRRIEERWLDEHPAAARQRPRGDTSS
MMAR_1245|M.marinum_M VAVVGLIWLAFEHRRVRRIADRWYSEHPEVRRQRL----AS
MUL_2642|M.ulcerans_Agy99 VAVVGLIWLAFEHRRVRRIADRWYSGHPEVRRQRL----AS
Mb3316c|M.bovis_AF2122/97 VTVVGLAWLAFEHRRIRKIADRWYTEHPEVRRQRL----AG
Rv3288c|M.tuberculosis_H37Rv VTVVGLAWLAFEHRRIRKIADRWYTEHPEVRRQRL----AG
MAV_4260|M.avium_104 AAVLGLVWLAVEHRRIRRIADRWYAEHPEVHRQWP----AS
MAB_2519c|M.abscessus_ATCC_199 FFFLGWLWMHVERRRVRRMEAEWHQEHSVPHHD--------
:. *: .*: *:*:: * *. ::