For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLGPLDEYPVHQIPQPIAWPGSSDRNFYDRSYFNAHDRTGNIFVISGIGYYPNLGVKDAFFLVRRGDTQT AVHLCDAIDQDRLNQHVNGYRIEVKEPLRKLRLVLDETEGVAADLTWEGLFDVVQEQPHILRSGNRVTLD AQRFAQLGSWSGRIVIDGQEIAVDPATWIGSRDRSWGIRPIGEPEPAGRPADPPFEGMWWLYVPMAFDDF AIVLIIQEQPDGFRSLNDCTRIWRDGRVEQLGWPRVKIHYRSGTRIPTGATIDASAPDGTPVHFDVESKL PVPIHVGGGYGGDSDWLHGMWKGEKFTERLTYDMTDPAIIARSGFGVIDHVGRAICRDGDKAPVEGWGLY EHGALGRHDPSGFSDWLTVAP
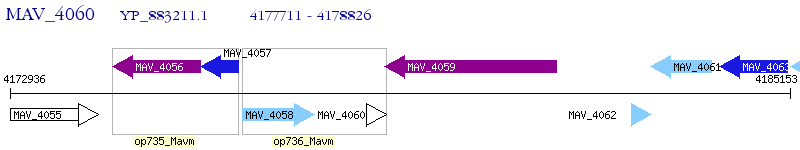
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4060 | - | - | 100% (371) | hypothetical protein MAV_4060 |
| M. avium 104 | MAV_5274 | - | 1e-11 | 24.59% (362) | hypothetical protein MAV_5274 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3194 | - | 0.0 | 88.14% (371) | hypothetical protein Mb3194 |
| M. gilvum PYR-GCK | Mflv_4498 | - | 0.0 | 84.37% (371) | hypothetical protein Mflv_4498 |
| M. tuberculosis H37Rv | Rv3169 | - | 0.0 | 88.14% (371) | hypothetical protein Rv3169 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4483 | - | 3e-65 | 38.24% (374) | hypothetical protein MAB_4483 |
| M. marinum M | MMAR_1392 | - | 0.0 | 92.70% (370) | hypothetical protein MMAR_1392 |
| M. smegmatis MC2 155 | MSMEG_4982 | - | 2e-31 | 29.56% (389) | hypothetical protein MSMEG_4982 |
| M. thermoresistible (build 8) | TH_0686 | - | 0.0 | 83.42% (380) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2484 | - | 0.0 | 92.70% (370) | hypothetical protein MUL_2484 |
| M. vanbaalenii PYR-1 | Mvan_1868 | - | 0.0 | 85.44% (371) | hypothetical protein Mvan_1868 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4498|M.gilvum_PYR-GCK ---MLGPMDEFPVHQIPQPIAWPGSSDRNFYDRSYFNAHDRTGDI-FVIT
Mvan_1868|M.vanbaalenii_PYR-1 ---MLGPMDEYPVHQVPQPIAWPGSSDRNFYDRAYYNAHDRTGDI-FVIT
MMAR_1392|M.marinum_M MPQPLGPLDEYPVHQLPQPIAWPGSSDRNFYDRSYFNAHDRTGNI-FVIS
MUL_2484|M.ulcerans_Agy99 MPQPLGPLDEYPVHQLPQPIAWPGSSDRNFYDRSYFNAHDRTGNI-FVIS
MAV_4060|M.avium_104 ---MLGPLDEYPVHQIPQPIAWPGSSDRNFYDRSYFNAHDRTGNI-FVIS
Mb3194|M.bovis_AF2122/97 MPQMLGPLDEYPLHQLPQPIAWPGSSDRNFYDRSYFNAHDRTGNI-FLIT
Rv3169|M.tuberculosis_H37Rv MPQMLGPLDEYPLHQLPQPIAWPGSSDRNFYDRSYFNAHDRTGNI-FLIT
TH_0686|M.thermoresistible__bu ---MLGPLDEFPVHQIPQPIAWPGSSDRNFYDRSYLNAHDRTGDI-FLIT
MAB_4483|M.abscessus_ATCC_1997 --MSLSPFDDYPIHQIADVVRHVGTSDRNFYDRYYFGCHGGRADLPYLIT
MSMEG_4982|M.smegmatis_MC2_155 ---MLTPHDELLCHQLPTTFDHVDQSDLRWTERIVMYGFDRSGDI-NVMT
* * *: **:. . . ** .: :* .. .:: :::
Mflv_4498|M.gilvum_PYR-GCK GLGYYPNLGVKDAFFLVRR---------GDTQTAVHLSDAIDQDR---LN
Mvan_1868|M.vanbaalenii_PYR-1 GIGYYPNLGVKDAFLLARR---------GDTQTAVHLSDAIDQDR---LN
MMAR_1392|M.marinum_M GIGYYPNLGVKDAFLLIRR---------GDTQTAVHLSDAIDQDR---LH
MUL_2484|M.ulcerans_Agy99 GIGYYPNLGVKDAFLLIRR---------GDTQTAVHLSDAIDQDR---LH
MAV_4060|M.avium_104 GIGYYPNLGVKDAFFLVRR---------GDTQTAVHLCDAIDQDR---LN
Mb3194|M.bovis_AF2122/97 GIGYYPNLGVKDAFVLIRR---------ADIQTAVHLSDAIDSDR---LH
Rv3169|M.tuberculosis_H37Rv GIGYYPNLGVKDAFVLIRR---------ADIQTAVHLSDAIDSDR---LH
TH_0686|M.thermoresistible__bu GIGYYPNLGVKDAFVLLARRGSGTSGGCGETQTAVHLSDAIDQDR---LN
MAB_4483|M.abscessus_ATCC_1997 GLGVYPNLGVTDAFASVRN---------GDDIVVFRASRALGDDR---AD
MSMEG_4982|M.smegmatis_MC2_155 GLARYPNRNVTDAYAMITRR--------DATARVVRMSTELNPETGPLGS
*:. *** .*.**: . ..: . :. :
Mflv_4498|M.gilvum_PYR-GCK QHVGNYRVEVIEPLRKLRIVLD-----ETDGIAADLTWEGLFDAVQEQPH
Mvan_1868|M.vanbaalenii_PYR-1 QHVGNYRVEVIEPLRKLRIVLD-----ETEGIAADLTWEGLFDVVQEQPH
MMAR_1392|M.marinum_M QHVNGYRIDVIEPLRKLRLVLD-----ETEGVSADLTWEGLFDVVAEQPH
MUL_2484|M.ulcerans_Agy99 QHVNGYRIDVIEPLRKLRLVLD-----ETEGVSADLTWEGLFDVVAEQPH
MAV_4060|M.avium_104 QHVNGYRIEVKEPLRKLRLVLD-----ETEGVAADLTWEGLFDVVQEQPH
Mb3194|M.bovis_AF2122/97 QHVNGYRVEVVEPLRKLRIVLD-----ETEGVAADLTWEGLFDVVQEQPH
Rv3169|M.tuberculosis_H37Rv QHVNGYRVEVVEPLRKLRIVLD-----ETEGVAADLTWEGLFDVVQEQPH
TH_0686|M.thermoresistible__bu QHVNGYRIEVLEPLRKLRVVLE-----ETEGMAVDLTWEGLFDPVQEQPH
MAB_4483|M.abscessus_ATCC_1997 LTVGPYRIEVVEGLKKVRVILEPGWHDATYDISFDLTYTGEIPASLEPGH
MSMEG_4982|M.smegmatis_MC2_155 YSVGPYTYTIEEPLRTVRAVLAP----NEQGVSLDLRLRGQFPTYEQTPA
*. * : * *:.:* :* .:: ** * : :
Mflv_4498|M.gilvum_PYR-GCK IMRRGTRVTLNAQRFAQVGSWSGQLSIDGTDIAVDPAQWIGTRDRSWGIR
Mvan_1868|M.vanbaalenii_PYR-1 IMRRGNRVTLDAQRFAQVGTWSGLLSIDGEDIAVDPARWIGTRDRSWGIR
MMAR_1392|M.marinum_M LLRSGNRVTLDAQRFAQLGTWSGQIQVDGQQIDVDPATWIGSRDRSWGIR
MUL_2484|M.ulcerans_Agy99 LLRSGNRVTLDAQRFAQLGTWSGQIQVDGQQIDVDPATWIGSRDRSWGIR
MAV_4060|M.avium_104 ILRSGNRVTLDAQRFAQLGSWSGRIVIDGQEIAVDPATWIGSRDRSWGIR
Mb3194|M.bovis_AF2122/97 VLRSGNRVTLDAQRFAQLGTWSGRIVVDGERIAVDPATWLGSRDRSWGIR
Rv3169|M.tuberculosis_H37Rv VLRSGNRVTLDAQRFAQLGTWSGRIVVDGERIAVDPATWLGSRDRSWGIR
TH_0686|M.thermoresistible__bu VLRSGNRVTLDAQRFAQVGTWSGVIVVDGEEIAVDPRHWIGTRDRSWGIR
MAB_4483|M.abscessus_ATCC_1997 YQRQLGRVMFDTSRFAQTGTWTGQLTIDGTTHELDGVNWSGNRDRSWGIR
MSMEG_4982|M.smegmatis_MC2_155 FHRSRGRVSEDARRFYQNGQVEGWIEIDGERIEIDPGSWWFGRDHSWGIR
* ** :: ** * * * : :** :* * **:*****
Mflv_4498|M.gilvum_PYR-GCK --PIGE--AEPAGRPDDPPFEGMWWLYIPMAFDDFGIVIIIQEDPLGFRS
Mvan_1868|M.vanbaalenii_PYR-1 --PVGE--AEPAGRPDDPPFEGMWWLYVPMAFDDFGIVIIIQEDPQGFRS
MMAR_1392|M.marinum_M --PVGE--PEPAGRPADPPFEGMWWLYVPMAFDDFAIVLIIQEEPSGFRS
MUL_2484|M.ulcerans_Agy99 --PVGE--PEPAGRPADPPFEGMWWLYVPMAFDDFAIVLIIQEEPSGFRS
MAV_4060|M.avium_104 --PIGE--PEPAGRPADPPFEGMWWLYVPMAFDDFAIVLIIQEQPDGFRS
Mb3194|M.bovis_AF2122/97 --PVGE--PEPAGRPADPPFEGMWWLYVPLAFDDFAVVLIIQEEPDGFRS
Rv3169|M.tuberculosis_H37Rv --PVGE--PEPAGRPADPPFEGMWWLYVPLAFDDFAVVLIIQEEPDGFRS
TH_0686|M.thermoresistible__bu --PIGE--PEPAGRPADPPFEGMWWLYVPMAFDDFAIVLIIQEEPDGFRS
MAB_4483|M.abscessus_ATCC_1997 --PVGE--GEPAGRRAT-LAGGFFWLYNVISFPQYSIVFINQEDEHGNKV
MSMEG_4982|M.smegmatis_MC2_155 NGPGGGSLSEGAHLQPQEIPDGTLYYMGIFQFDDELVHFAQRETSTGERW
* * * * * : : * : : : :* * :
Mflv_4498|M.gilvum_PYR-GCK LNDCTRVWKDGR---VEQLGWPRVKIQYRSGTRIPTGATVEATALDGSPL
Mvan_1868|M.vanbaalenii_PYR-1 LNDCTRIWKDGR---VEQLGWPRVKIHYRSGTRIPTGATIEATAFDGSPL
MMAR_1392|M.marinum_M LNDCTRIWRDGR---VEQLGWPRVKIHYRSGTRIPTGATIDATRPDGTPV
MUL_2484|M.ulcerans_Agy99 LNDCTRIWRDGR---VEQLGWPRVKIHYRSGTRIPTGATIDATRPDGTPV
MAV_4060|M.avium_104 LNDCTRIWRDGR---VEQLGWPRVKIHYRSGTRIPTGATIDASAPDGTPV
Mb3194|M.bovis_AF2122/97 LNDCTRIWRDGH---VEQLGWPRVRIHYRSGTRIPTGATIEASTPDGAPV
Rv3169|M.tuberculosis_H37Rv LNDCTRIWRDGH---VEQLGWPRVRIHYRSGTRIPTGATIEASTPDGAPV
TH_0686|M.thermoresistible__bu LNDCTRIFRDGR---VEQLGWPRVQIHYTSGTRIPYGATITASTPDGTPL
MAB_4483|M.abscessus_ATCC_1997 VSGARRVWRDPSK-GIEELGPIAHDITLIPGTREARSAVVTLG-----DI
MSMEG_4982|M.smegmatis_MC2_155 QFEGEVLYPIETGRPAGTIIDVEHDLKFRDDLRVISGGTFTVHRGDRTTS
:: : : . * ....
Mflv_4498|M.gilvum_PYR-GCK HFDVESKLAVPIHVGGGYGGDSDWLHGVWKGPNFTERLTYDMTDPAVIGR
Mvan_1868|M.vanbaalenii_PYR-1 RFEVESKLPVPIHVGGGYGGDSDWLHGVWKGEGFTERLTYDMTDPAIVGR
MMAR_1392|M.marinum_M HFDVESKLAVPIHVGGGYGGDSDWLHGMWKGEKFVERLTYGMTDPAIIAR
MUL_2484|M.ulcerans_Agy99 HFDVESKLAVPIHVGGGYGGDSDWLHGMWKGEKFVERLTYGMTDPAIIAR
MAV_4060|M.avium_104 HFDVESKLPVPIHVGGGYGGDSDWLHGMWKGEKFTERLTYDMTDPAIIAR
Mb3194|M.bovis_AF2122/97 HFDVESKLAVPTHVGGGYGGDSDWSHGMWKGEKFVERRTYDMTDPTIIAR
Rv3169|M.tuberculosis_H37Rv HFDVESKLAVPTHVGGGYGGDSDWSHGMWKGEKFVERRTYDMTDPTIIAR
TH_0686|M.thermoresistible__bu VFEVESKLAVPIHIGGGYGGDSDWLHGMWKGEKFTERLTYDMTDPSVSGR
MAB_4483|M.abscessus_ATCC_1997 TITADIVLPHYFGFGTGYGLDPDWRHGMWQGDLVVQGKRYKTAELDATLK
MSMEG_4982|M.smegmatis_MC2_155 TIEVRPMTDFWPGLAG-YMEVDGYASGHWRGRDFIDGFTADIGDIDVVKP
: . .. * .: * *:* . : :
Mflv_4498|M.gilvum_PYR-GCK AGFGVIDHVGRAVCREADGTEREGWGLFEHGALGRHDPSGFADWLTVAP-
Mvan_1868|M.vanbaalenii_PYR-1 AGFGVIDHVGRAVCRDADGTEREGWGLFEHGALGRHDPSGFADWLTVAP-
MMAR_1392|M.marinum_M SGFGVIDHVGRAVCRDGDTDPVEGWGLYEHGALGRHDPSGFSDWLTVAP-
MUL_2484|M.ulcerans_Agy99 SGFGVIDHVGRAVCRDGDTDPVEGWGLYEHGALGRHDPSGFSDWLTVAP-
MAV_4060|M.avium_104 SGFGVIDHVGRAICRDGDKAPVEGWGLYEHGALGRHDPSGFSDWLTVAP-
Mb3194|M.bovis_AF2122/97 AGFGVIDHVGRALCRDGDGNPVQGWGLFEHGALGRHDPSGFADWSTLAP-
Rv3169|M.tuberculosis_H37Rv AGFGVIDHVGRALCRDGDGNPVQGWGLFEHGALGRHDPSGFADWSTLAP-
TH_0686|M.thermoresistible__bu AMFGVIDHVGRATCH-IDGRTVEGWGLFEHGALGRHDPSGFTDWMSVAP-
MAB_4483|M.abscessus_ATCC_1997 L-LCPVDNLA-TFRLIENGVETHGSGLFEFGPIGPHKPSGFTGFVDTHQE
MSMEG_4982|M.smegmatis_MC2_155 V--SFLSETLCEVSMDSGSGPKIGHGLVEMVFVGAYPRYGYTGW------
:.. . * ** * :* : *::.:
Mflv_4498|M.gilvum_PYR-GCK -----
Mvan_1868|M.vanbaalenii_PYR-1 -----
MMAR_1392|M.marinum_M -----
MUL_2484|M.ulcerans_Agy99 -----
MAV_4060|M.avium_104 -----
Mb3194|M.bovis_AF2122/97 -----
Rv3169|M.tuberculosis_H37Rv -----
TH_0686|M.thermoresistible__bu -----
MAB_4483|M.abscessus_ATCC_1997 RDSDS
MSMEG_4982|M.smegmatis_MC2_155 -----