For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KFTLNILEKRGNDDDTEAEHRWPQYVFGGRMRTSTFVLIVAFLLVWWVYDTYRPEPAPKPPAQQVVPPGF VPDPNYTWVPRSRLQQPPATVTYTPTPTSTPPPPPPPPPVTTTTTTTTTPPFQLPKLPCILPAPFCPPST TPSTPAPEQPQPGPGPTPTSPPPAS*
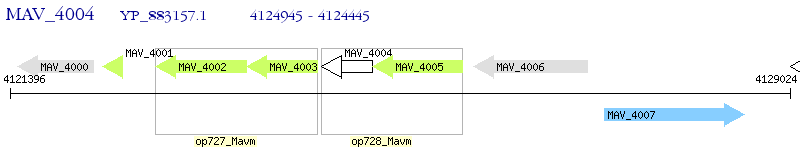
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4004 | - | - | 100% (166) | hypothetical protein MAV_4004 |
| M. avium 104 | MAV_4848 | - | 1e-09 | 29.38% (160) | hypothetical protein MAV_4848 |
| M. avium 104 | MAV_4606 | - | 5e-09 | 32.56% (129) | hypothetical protein MAV_4606 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3130c | - | 3e-41 | 57.89% (152) | hypothetical protein Mb3130c |
| M. gilvum PYR-GCK | Mflv_4449 | - | 9e-30 | 45.73% (164) | hypothetical protein Mflv_4449 |
| M. tuberculosis H37Rv | Rv3103c | - | 3e-41 | 57.89% (152) | hypothetical protein Rv3103c |
| M. leprae Br4923 | MLBr_00877 | mmpS3 | 4e-07 | 31.09% (119) | hypothetical protein MLBr_00877 |
| M. abscessus ATCC 19977 | MAB_3476c | - | 1e-19 | 40.56% (143) | hypothetical protein MAB_3476c |
| M. marinum M | MMAR_1529 | - | 1e-42 | 53.29% (167) | hypothetical protein MMAR_1529 |
| M. smegmatis MC2 155 | MSMEG_2088 | - | 2e-27 | 45.39% (141) | hypothetical protein MSMEG_2088 |
| M. thermoresistible (build 8) | TH_2661 | - | 2e-27 | 43.37% (166) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2410 | - | 4e-42 | 52.69% (167) | hypothetical protein MUL_2410 |
| M. vanbaalenii PYR-1 | Mvan_1913 | - | 4e-34 | 49.37% (158) | hypothetical protein Mvan_1913 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3130c|M.bovis_AF2122/97 ----------------------------------MKLS------------
Rv3103c|M.tuberculosis_H37Rv ----------------------------------MKLS------------
MMAR_1529|M.marinum_M ----------------------------------MKITFVPTRLRRSGQS
MUL_2410|M.ulcerans_Agy99 ----------------------------------MKITFVPTRLRRSGQS
MAV_4004|M.avium_104 ----------------------------------MKFTLN-ILEKRGNDD
Mflv_4449|M.gilvum_PYR-GCK -----------------------------------------MTLPSFLQR
Mvan_1913|M.vanbaalenii_PYR-1 --------------------------------------------MRFIRR
TH_2661|M.thermoresistible__bu --------------------------------------------------
MSMEG_2088|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3476c|M.abscessus_ATCC_199 -----------------------------------------MSEPRHEPF
MLBr_00877|M.leprae_Br4923 MSGPNPPGRENEESDSGNELSGELDPHNGVESVDELVPVPDSDLVTASDH
Mb3130c|M.bovis_AF2122/97 --NQKRHWPGYLFG-RIRTSTLVLIAAFLAVWWIYETYRPQAPGPGDSPP
Rv3103c|M.tuberculosis_H37Rv --NQKRHWPGYLFG-RIRTSTLVLIAAFLAVWWIYETYRPQAPGPGDSPP
MMAR_1529|M.marinum_M NGNRKRGTPRQLFGGRIRLSTVVLMVAFLAVWWLYDTYNPQHSAGKTTPP
MUL_2410|M.ulcerans_Agy99 NGNRKRGTPRQLFGGRIRLSTVVLMVAFLAVWWLYDTYNPQHSAGKTTPP
MAV_4004|M.avium_104 DTEAEHRWPQYVFGGRMRTSTFVLIVAFLLVWWVYDTYRPE-PAPK-PPA
Mflv_4449|M.gilvum_PYR-GCK LQPKNRHSRAYLFGGRMRVSTVGLVLVFFALYWVNQNYQPEPPAPAMDPA
Mvan_1913|M.vanbaalenii_PYR-1 RDGESRGWPTYLLGGRIRASTAGLILAFLALFWVNQNYQPELPAPTPDPA
TH_2661|M.thermoresistible__bu MPDRRLRIPGRLFGGRIRTSTVALIAAFIGLVWLDQVYEP---PPPPEPV
MSMEG_2088|M.smegmatis_MC2_155 ---------------------MVLIVAFFALWWLQQTYQP---EPARTET
MAB_3476c|M.abscessus_ATCC_199 WEMALSRAKKLRKKIRFRTSTVVLVLAFIATSWLYDITRPEPAPPQPPPG
MLBr_00877|M.leprae_Br4923 TSETEVYSQAYSAPEAEHFTAVPYVPADLRLYDYDESSVYDEPGAAPRWP
: . : :
Mb3130c|M.bovis_AF2122/97 TQVVPPGFVPDPDYTWVPRTRVQPP--TVKATPTTTSSTP---------P
Rv3103c|M.tuberculosis_H37Rv TQVVPPGFVPDPDYTWVPRTRVQPP--TVKATPTTTSSTP---------P
MMAR_1529|M.marinum_M SQVVPPGFVPDPNYTWVPRTRVQPP--TTATRPTPTTTSPT--------P
MUL_2410|M.ulcerans_Agy99 SQVVPPGLVPDPNYTWVPRTRVQPP--TTATRPTPTTTSPT--------P
MAV_4004|M.avium_104 QQVVPPGFVPDPNYTWVPRSRLQQPPATVTYTPTPTSTPPP--------P
Mflv_4449|M.gilvum_PYR-GCK QQVVPPGFVPDPNYTWVPRTNVETRAPEPTTTTTTTTTTPT------TTT
Mvan_1913|M.vanbaalenii_PYR-1 QQVVPPGFVPDPNYTWVPRTNVAPRQPEVTTTTPTTTTT---------TT
TH_2661|M.thermoresistible__bu QQWVPPGFVPNPDYTWVPRT--AVRRPPTTESEPTTTTT---------TT
MSMEG_2088|M.smegmatis_MC2_155 PQVVPPGFVPDPDYTWVPRTKVETPRTTRTTTPTTTTTTPPPTSETPTTT
MAB_3476c|M.abscessus_ATCC_199 YTWVPKPSVTTTTQQPVPTTTRRRTTTTPPTTTTTTTEQTN------TIV
MLBr_00877|M.leprae_Br4923 WVVGVAAILAAISLVVSVSLLFTRTDTSKLSTPTTGRSTPPVQDEITTVK
:. ..
Mb3130c|M.bovis_AF2122/97 VSPPETTTDSAVP--PPFELP----------------PPFGPGTTTPTPP
Rv3103c|M.tuberculosis_H37Rv VSPPETTTDSAVP--PPFELP----------------PPFGPGTTTPTPP
MMAR_1529|M.marinum_M TPTPTTSTPVPGPSAPPCLLP----------------PPFCPATTTPPTP
MUL_2410|M.ulcerans_Agy99 TPTPTTSTPVPGPSAPPCLLP----------------PPFCPATTTPPTP
MAV_4004|M.avium_104 PPPPPVTTTTTTTTTPPFQLPK---------LPCILPAPFCPPSTTPSTP
Mflv_4449|M.gilvum_PYR-GCK TTTPSPTPDTPTTSPETSTPP-PAGPLFPSEPTTTVIDPDGPGPLAPQTF
Mvan_1913|M.vanbaalenii_PYR-1 TTPPETTTATTTAEPTPSTTPGPLGPVVPFGPTTTVIDPDGPGPLSPQTF
TH_2661|M.thermoresistible__bu TTPP---ADTPTTTPPAVTTP--------EEPTTTVIDPDGPGLLPPTTV
MSMEG_2088|M.smegmatis_MC2_155 PTGPLGPGDTTSPTSPTSTSP---------GPATTVVDPDGPGILPPITL
MAB_3476c|M.abscessus_ATCC_199 TTFPTLTPSGTAPTPPTTTTP------------------APPGIVPPWLV
MLBr_00877|M.leprae_Br4923 PPPPSTETSTATETQTVTVTPLPPPSATSTAVPPSSVVPPPPTTPTTTVT
. * . * * ..
Mb3130c|M.bovis_AF2122/97 AP--------LPQPG--PGPTAG----TYPKSEPPTR-------------
Rv3103c|M.tuberculosis_H37Rv AP--------LPQPG--PGPTAG----TYPKSEPPTR-------------
MMAR_1529|M.marinum_M VPSPS----QTPEPE--PGPVS-----SGPTPTPPAS-------------
MUL_2410|M.ulcerans_Agy99 VPSPS----QTPEPE--PGPVS-----SGPTPTPPAS-------------
MAV_4004|M.avium_104 AP-------EQPQPG--PGPTP--------TSPPPAS-------------
Mflv_4449|M.gilvum_PYR-GCK TQ-------APPVLT--PTTVAP----PGPVLPPTPP---QP--------
Mvan_1913|M.vanbaalenii_PYR-1 TQ-------APPVP---PTTVTA----PAPTPTAPPP---LP--------
TH_2661|M.thermoresistible__bu TETDRPDLGAPPTPWGPPTTTTAPAFGPGQQPQNQPPGAARPPGG-----
MSMEG_2088|M.smegmatis_MC2_155 PVLPG----AAPAPA--PTPTPE----PATEPAAPAP----PQ-------
MAB_3476c|M.abscessus_ATCC_199 PP-------QLPQLPPMPGAPPP----PGQNPAP----------------
MLBr_00877|M.leprae_Br4923 TLTGPRQVTYSVTGTKAPGDIISVTYVDASGRRRTQHNVYIPWSMTVTPI
*
Mb3130c|M.bovis_AF2122/97 -------------------------------------------
Rv3103c|M.tuberculosis_H37Rv -------------------------------------------
MMAR_1529|M.marinum_M -------------------------------------------
MUL_2410|M.ulcerans_Agy99 -------------------------------------------
MAV_4004|M.avium_104 -------------------------------------------
Mflv_4449|M.gilvum_PYR-GCK -------------------------------------------
Mvan_1913|M.vanbaalenii_PYR-1 -------------------------------------------
TH_2661|M.thermoresistible__bu -------------------------------------------
MSMEG_2088|M.smegmatis_MC2_155 -------------------------------------------
MAB_3476c|M.abscessus_ATCC_199 -------------------------------------------
MLBr_00877|M.leprae_Br4923 SQSDVGSVEAFSLFRVSKLNCLITTSDGTVLSSNSNDAPQTSC