For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIASVVARQLLDCKARPLVEVEITTDTGHVGRGAAPTGTSVGAHEAFVLRDGEPTRYRGRSVHRAVAAVR DEIAPVLTGAELDDPRSLDRVMIELDGTPDKHRLGGNAIYSTSIALLRAAAAAAGTPTYTYVGALLGLTP PTTVPMPSFNMINGGRYGDVEQSFSEFLVVPYRAESIQAAVEKGVSLFEVLGEVLAEHLGRTPLLASSYG YIAPSGDPHAVLELLAEAVERAGCADVMAFALDCASSEVYDNGSATYAFNGGRVTAEALIDYARALSQEF PMLFIEDLLDGDDWAGFTKAVQTVNRSIIVGDDLIVTNRDRLRRAVETSAVDGFILKPNQVGTIAEALDC FEYASQNGLLAIPSGRSGGVIDDVVMDLAVGLGAPLQKNGAPRSGERIEKLNFLLRAAEGIPDCALADVP ALARF*
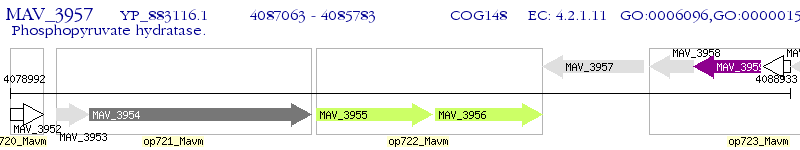
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3957 | - | - | 100% (426) | enolase |
| M. avium 104 | MAV_1164 | eno | 6e-76 | 39.09% (417) | phosphopyruvate hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1051 | eno | 5e-76 | 39.47% (418) | phosphopyruvate hydratase |
| M. gilvum PYR-GCK | Mflv_1954 | eno | 1e-73 | 39.71% (418) | phosphopyruvate hydratase |
| M. tuberculosis H37Rv | Rv1023 | eno | 5e-76 | 39.47% (418) | phosphopyruvate hydratase |
| M. leprae Br4923 | MLBr_00255 | eno | 8e-73 | 38.52% (418) | phosphopyruvate hydratase |
| M. abscessus ATCC 19977 | MAB_1165 | - | 2e-73 | 38.76% (418) | enolase (Eno) |
| M. marinum M | MMAR_4462 | eno | 6e-73 | 38.28% (418) | enolase Eno |
| M. smegmatis MC2 155 | MSMEG_5415 | eno | 3e-76 | 39.33% (417) | phosphopyruvate hydratase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4631 | eno | 5e-73 | 38.28% (418) | phosphopyruvate hydratase |
| M. vanbaalenii PYR-1 | Mvan_4771 | eno | 2e-74 | 39.71% (418) | phosphopyruvate hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1165|M.abscessus_ATCC_1997 ------------------MPILEQVGAREILDSRGNPTVEVEVALTDGTF
MSMEG_5415|M.smegmatis_MC2_155 ------------------MPIIEQVGAREILDSRGNPTVEVEVALTDGTF
Mflv_1954|M.gilvum_PYR-GCK ------------------MPIIEQVGAREILDSRGNPTVEVEVGLLDGTV
Mvan_4771|M.vanbaalenii_PYR-1 ------------------MPIIEQVGAREILDSRGNPTVEVEVGLLDGTV
Mb1051|M.bovis_AF2122/97 ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
Rv1023|M.tuberculosis_H37Rv ------------------MPIIEQVRAREILDSRGNPTVEVEVALIDGTF
MMAR_4462|M.marinum_M ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
MUL_4631|M.ulcerans_Agy99 ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
MLBr_00255|M.leprae_Br4923 MSPPTGRPRLSHDYKENLVPVIEQVGAREILDSRGNPTVEVEVVLIDGTF
MAV_3957|M.avium_104 -------------------MRIASVVARQLLDCKARPLVEVEITTDTGHV
: .* **::**.:..* ****: * .
MAB_1165|M.abscessus_ATCC_1997 ARAAVPSGASTGEHEAVELRDGD-ARYGGKGVTKAVNAVLDEIAPAIIGE
MSMEG_5415|M.smegmatis_MC2_155 ARAAVPSGASTGEHEAVELRDGG-SRYGGKGVEKAVEAVLDEIAPQVIGL
Mflv_1954|M.gilvum_PYR-GCK ARAAVPSGASTGEHEAVELRDGG-SRYLGKGVEKAVEAVLDEIAPAVIGL
Mvan_4771|M.vanbaalenii_PYR-1 SRAAVPSGASTGEHEAVELRDGG-SRYLGKGVEKAVEAVLDEIAPAVIGL
Mb1051|M.bovis_AF2122/97 ARAAVPSGASTGEHEAVELRDGG-DRYGGKGVQKAVQAVLDEIGPAVIGL
Rv1023|M.tuberculosis_H37Rv ARAAVPSGASTGEHEAVELRDGG-DRYGGKGVQKAVQAVLDEIGPAVIGL
MMAR_4462|M.marinum_M ARAAVPSGASTGEHEAVELRDGG-DRYGGKGVKKAVEAVLDEIGPAVIGL
MUL_4631|M.ulcerans_Agy99 ARAAVPSGASTGEHEAVELRDGG-DRYGGKGVKKAVEAVLDEIGPAVIGL
MLBr_00255|M.leprae_Br4923 ARAAVPSGASTGEYEAVELRDGD-GRYGGKGVKRAVDAVLDEIGPVVIGL
MAV_3957|M.avium_104 GRGAAPTGTSVGAHEAFVLRDGEPTRYRGRSVHRAVAAVRDEIAPVLTGA
.*.*.*:*:*.* :**. **** ** *:.* :** ** ***.* : *
MAB_1165|M.abscessus_ATCC_1997 SADDQRLIDQALLDLDGTPDKSRLGANAILGVSLAVAKAAADSAGLALFR
MSMEG_5415|M.smegmatis_MC2_155 SADDQRLVDQALLDLDGTPDKSRLGANAILGVSLAVSKAAAESAGLPLFR
Mflv_1954|M.gilvum_PYR-GCK GADEQRLVDQALLDLDGTPDKSRLGANAILGVSLAVAKAAAQSAELPLFR
Mvan_4771|M.vanbaalenii_PYR-1 GADEQRLVDQALVDLDGTPDKSRLGANAILGVSLAVAKAAAQSAELPLFR
Mb1051|M.bovis_AF2122/97 NADDQRLVDQALVDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFR
Rv1023|M.tuberculosis_H37Rv NADDQRLVDQALVDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFR
MMAR_4462|M.marinum_M NADDQRLVDQALVDLDGTPDKSRLGGNSILGVSLAVAKAASESAELPLFR
MUL_4631|M.ulcerans_Agy99 NADDQRLVDQALVDLDGTPDKSRLGGNSILGVSLAVAKAASESAELPLFR
MLBr_00255|M.leprae_Br4923 NANDQRLIDQELLDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFR
MAV_3957|M.avium_104 ELDDPRSLDRVMIELDGTPDKHRLGGNAIYSTSIALLRAAAAAAGTPTYT
:: * :*: :::******* ***.*:* ..*:*: :**: :* . :
MAB_1165|M.abscessus_ATCC_1997 YLGGPN----AHILPVPMMNILNGGAHADTGVDVQEFMVAPIGAPSFKES
MSMEG_5415|M.smegmatis_MC2_155 YIGGPN----AHILPVPMMNILNGGAHADTGVDVQEFMVAPIGAPSFKEA
Mflv_1954|M.gilvum_PYR-GCK YLGGPN----AHILPVPMMNIINGGAHADTGVDVQEFMIAPIGAPSFKEA
Mvan_4771|M.vanbaalenii_PYR-1 YVGGPN----AHILPVPMMNIINGGAHADTGVDVQEFMIAPIGAPSFKEA
Mb1051|M.bovis_AF2122/97 YVGGPN----AHILPVPMMNILNGGAHADTAVDIQEFMVAPIGAPSFVEA
Rv1023|M.tuberculosis_H37Rv YVGGPN----AHILPVPMMNILNGGAHADTAVDIQEFMVAPIGAPSFVEA
MMAR_4462|M.marinum_M YIGGPN----AHILPVPMMNILNGGAHADTGVDIQEFMVAPIGAPSFSEA
MUL_4631|M.ulcerans_Agy99 YIGGPN----AHILPVPMMNILNGGAHADTGVDIQEFMVAPIGAPSFSEA
MLBr_00255|M.leprae_Br4923 YIGGSN----AHILPVPMMNILNGGAHADTAVDVQEFMVAPIGAPSFVEA
MAV_3957|M.avium_104 YVGALLGLTPPTTVPMPSFNMINGGRYGDVEQSFSEFLVVPYRAESIQAA
*:*. . :*:* :*::*** :.*. ...**::.* * *: :
MAB_1165|M.abscessus_ATCC_1997 LRWGTEVYHSLKSVLKKQ-GLSTGLGDEGGFAPDVAGTRAALDLISTAIE
MSMEG_5415|M.smegmatis_MC2_155 LRWGAEVYHSLKSVLKNQ-GLATGLGDEGGFAPDVAGTKAALDLISSAIE
Mflv_1954|M.gilvum_PYR-GCK LRWGAEVYHSLKSVLKKQ-GLSTGLGDEGGFAPDLPGTKAALDLIGTAIE
Mvan_4771|M.vanbaalenii_PYR-1 LRWGAEVYHSLKSVLKKQ-GLATGLGDEGGFAPDLPGTRAALDLIGTAIE
Mb1051|M.bovis_AF2122/97 LRWGAEVYHALKSVLKKE-GLSTGLGDEGGFAPDVAGTTAALDLISRAIE
Rv1023|M.tuberculosis_H37Rv LRWGAEVYHALKSVLKKE-GLSTGLGDEGGFAPDVAGTTAALDLISRAIE
MMAR_4462|M.marinum_M LRWGAEVYHALKAVLKKA-GLSTGLGDEGGFAPDVASTTAALDLISQAIE
MUL_4631|M.ulcerans_Agy99 LRWGAEVYHALKAVLKKA-GLSTGLGDEGGFAPDVASTTAALDLISQAIE
MLBr_00255|M.leprae_Br4923 LRWGAEVYHALKSVLKKK-GLSTGLGDEGGFAPEVAGTTAALDLVTLAIE
MAV_3957|M.avium_104 VEKGVSLFEVLGEVLAEHLGRTPLLASSYGYIAPSGDPHAVLELLAEAVE
:. *..::. * ** : * :. *... *: . .. *.*:*: *:*
MAB_1165|M.abscessus_ATCC_1997 ATGLKLGADVALALDVAATEFYSAADG-YSFEKEKRTAEQMGAFYAELLD
MSMEG_5415|M.smegmatis_MC2_155 ATGLKLGSDVALALDVAATEFYTEGSG-YAFEKETRTAEQMAEFYAGLLD
Mflv_1954|M.gilvum_PYR-GCK GAGLKIGSDVALALDVAATEFYTDGTG-YAFEKETRTAEQMAQFYEQLIG
Mvan_4771|M.vanbaalenii_PYR-1 AAGLKVGSDVALALDVAATEFYTEGTG-YAFEKETRTAEQMAAFYAQLLE
Mb1051|M.bovis_AF2122/97 SAGLRPGADVALALDAAATEFFTDGTG-YVFEGTTRTADQMTEFYAGLLG
Rv1023|M.tuberculosis_H37Rv SAGLRPGADVALALDAAATEFFTDGTG-YVFEGTTRTADQMTEFYAGLLG
MMAR_4462|M.marinum_M AAGFKPGVDVALALDAAANEFHADGS--YTFEGTPRTAAQMTEFYAGLLG
MUL_4631|M.ulcerans_Agy99 AAGFKPGVDVALALDAAANEFHADGS--YTFEGTPRTAAQMTEFYAGLLG
MLBr_00255|M.leprae_Br4923 AAGFKPGADVALALDAAATEFYTDGIG-YHFEGMTHTADQMTEFYADLLG
MAV_3957|M.avium_104 RAGCAD--VMAFALDCASSEVYDNGSATYAFNGGRVTAEALIDYARALSQ
:* :*:*** *:.*.. . * *: ** : : *
MAB_1165|M.abscessus_ATCC_1997 AYPLVSIEDPLSEDDWDGWVALTTAIGDRVQLVGDDLFVTNPERLEEGIE
MSMEG_5415|M.smegmatis_MC2_155 SYPLVSIEDPLSEDDWDGWVSLTAAIGDRIQLVGDDLFVTNPERLEDGIQ
Mflv_1954|M.gilvum_PYR-GCK AYPLVSIEDPLSEDDWDGWVALTSAIGDRVQLVGDDLFVTNPERLEDGIE
Mvan_4771|M.vanbaalenii_PYR-1 AYPLVSIEDPLSEDDWDGWVALTAAIGDKVQLVGDDLFVTNPERLEDGIE
Mb1051|M.bovis_AF2122/97 AYPLVSIEDPLSEDDWDGWAALTASIGDRVQIVGDDIFVTNPERLEEGIE
Rv1023|M.tuberculosis_H37Rv AYPLVSIEDPLSEDDWDGWAALTASIGDRVQIVGDDIFVTNPERLEEGIE
MMAR_4462|M.marinum_M SYPLVSIEDPLYENDWDGWAALTAEIGDRVQIVGDDVFVTNPERLEEGID
MUL_4631|M.ulcerans_Agy99 SYPVVSIEDPLYENDWDGWAALTAEIGDRVQIVGDDVFVTNPERLEEGID
MLBr_00255|M.leprae_Br4923 SYPLVSIEDPLSEDDWDGWAALTASIGEQVQIVGDDIFATNPERLEEGIG
MAV_3957|M.avium_104 EFPMLFIEDLLDGDDWAGFTKAVQTVN-RSIIVGDDLIVTNRDRLRRAVE
:*:: *** * :** *:. . :. : :****::.** :**. .:
MAB_1165|M.abscessus_ATCC_1997 RGAANALLVKVNQIGTLTETLDAVTLAHSSGYKTMMSHRSGETEDTTIAD
MSMEG_5415|M.smegmatis_MC2_155 RGAANALLVKVNQIGTLTETLDAVSLAHNSGYRTMMSHRSGETEDTTIAD
Mflv_1954|M.gilvum_PYR-GCK RGAGNALLVKVNQIGTLTETLDAVSLAHHAGYKTMMSHRSGETEDTTIAD
Mvan_4771|M.vanbaalenii_PYR-1 RGAANALLVKVNQIGTLTETLDAVSLAHNAGYKTMMSHRSGETEDTTIAD
Mb1051|M.bovis_AF2122/97 RGVANALLVKVNQIGTLTETLDAVTLAHHGGYRTMISHRSGETEDTMIAD
Rv1023|M.tuberculosis_H37Rv RGVANALLVKVNQIGTLTETLDAVTLAHHGGYRTMISHRSGETEDTMIAD
MMAR_4462|M.marinum_M RGVANALLVKVNQIGTLTETLDAVALAHHSGYRTMISHRSGETEDTIIAD
MUL_4631|M.ulcerans_Agy99 RGVANALLVKVNQIGTLTETLDAVALAHHSGYRTMISHRSGETEDTIIAD
MLBr_00255|M.leprae_Br4923 RGVANALLVKVNQIGTLTETLEAVALAHHSGYRTMISHRSGETEDTMIAD
MAV_3957|M.avium_104 TSAVDGFILKPNQVGTIAEALDCFEYASQNGLLAIPSGRSGGVIDDVVMD
.. :.:::* **:**::*:*:.. * * :: * *** . * : *
MAB_1165|M.abscessus_ATCC_1997 LAVAVGSGQIKTGAPARSERVAKYNQLLRIEETLGDAARFAGDLAFPRFS
MSMEG_5415|M.smegmatis_MC2_155 LAVAVGSGQIKTGAPARSERVAKYNQLLRIEETLGDAARYAGDLAFPRLE
Mflv_1954|M.gilvum_PYR-GCK LAVAVGSGQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFPRFG
Mvan_4771|M.vanbaalenii_PYR-1 LAVAVGSGQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFPRFS
Mb1051|M.bovis_AF2122/97 LAVAIGSGQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFA
Rv1023|M.tuberculosis_H37Rv LAVAIGSGQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFA
MMAR_4462|M.marinum_M LAVAVGSGQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFV
MUL_4631|M.ulcerans_Agy99 LAVAVGSGQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFV
MLBr_00255|M.leprae_Br4923 LVVALGSGQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFLRYV
MAV_3957|M.avium_104 LAVGLGAPLQKNGAPRSGERIEKLNFLLRAAEGIPDCA-LADVPALARF-
*.*.:*: *.*** .**: * * *** * : *.* *. *: *
MAB_1165|M.abscessus_ATCC_1997 IEPAN
MSMEG_5415|M.smegmatis_MC2_155 AK---
Mflv_1954|M.gilvum_PYR-GCK VESK-
Mvan_4771|M.vanbaalenii_PYR-1 VETK-
Mb1051|M.bovis_AF2122/97 CETK-
Rv1023|M.tuberculosis_H37Rv CETK-
MMAR_4462|M.marinum_M ADPK-
MUL_4631|M.ulcerans_Agy99 ADPK-
MLBr_00255|M.leprae_Br4923 VETR-
MAV_3957|M.avium_104 -----