For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PIIEQVGAREILDSRGNPTVEVEVALIDGTFARAAVPSGASTGEHEAVELRDGGDRYGGKGVKKAVEAVL DEIGPAVIGLNADDQRLVDQALVDLDGTPDKSRLGGNSILGVSLAVAKAASESAELPLFRYIGGPNAHIL PVPMMNILNGGAHADTGVDIQEFMVAPIGAPSFSEALRWGAEVYHALKAVLKKAGLSTGLGDEGGFAPDV ASTTAALDLISQAIEAAGFKPGVDVALALDAAANEFHADGSYTFEGTPRTAAQMTEFYAGLLGSYPVVSI EDPLYENDWDGWAALTAEIGDRVQIVGDDVFVTNPERLEEGIDRGVANALLVKVNQIGTLTETLDAVALA HHSGYRTMISHRSGETEDTIIADLAVAVGSGQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFP RFVADPK*
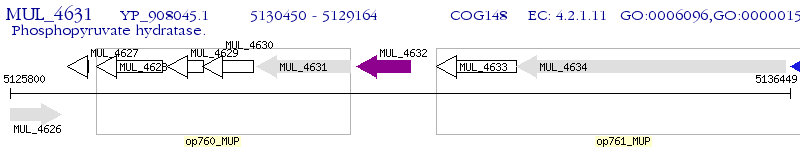
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4631 | eno | - | 100% (428) | phosphopyruvate hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1051 | eno | 0.0 | 90.91% (429) | phosphopyruvate hydratase |
| M. gilvum PYR-GCK | Mflv_1954 | eno | 0.0 | 83.45% (429) | phosphopyruvate hydratase |
| M. tuberculosis H37Rv | Rv1023 | eno | 0.0 | 90.68% (429) | phosphopyruvate hydratase |
| M. leprae Br4923 | MLBr_00255 | eno | 0.0 | 86.48% (429) | phosphopyruvate hydratase |
| M. abscessus ATCC 19977 | MAB_1165 | - | 0.0 | 83.92% (429) | enolase (Eno) |
| M. marinum M | MMAR_4462 | eno | 0.0 | 99.77% (428) | enolase Eno |
| M. avium 104 | MAV_1164 | eno | 0.0 | 89.51% (429) | phosphopyruvate hydratase |
| M. smegmatis MC2 155 | MSMEG_5415 | eno | 0.0 | 86.38% (426) | phosphopyruvate hydratase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4771 | eno | 0.0 | 83.45% (429) | phosphopyruvate hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1051|M.bovis_AF2122/97 ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
Rv1023|M.tuberculosis_H37Rv ------------------MPIIEQVRAREILDSRGNPTVEVEVALIDGTF
MUL_4631|M.ulcerans_Agy99 ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
MMAR_4462|M.marinum_M ------------------MPIIEQVGAREILDSRGNPTVEVEVALIDGTF
MLBr_00255|M.leprae_Br4923 MSPPTGRPRLSHDYKENLVPVIEQVGAREILDSRGNPTVEVEVVLIDGTF
MAV_1164|M.avium_104 ------------------MPIIEQVGAREILDSRGNPTVEVEIALTDGTF
Mflv_1954|M.gilvum_PYR-GCK ------------------MPIIEQVGAREILDSRGNPTVEVEVGLLDGTV
Mvan_4771|M.vanbaalenii_PYR-1 ------------------MPIIEQVGAREILDSRGNPTVEVEVGLLDGTV
MSMEG_5415|M.smegmatis_MC2_155 ------------------MPIIEQVGAREILDSRGNPTVEVEVALTDGTF
MAB_1165|M.abscessus_ATCC_1997 ------------------MPILEQVGAREILDSRGNPTVEVEVALTDGTF
:*::*** ****************: * ***.
Mb1051|M.bovis_AF2122/97 ARAAVPSGASTGEHEAVELRDGGDRYGGKGVQKAVQAVLDEIGPAVIGLN
Rv1023|M.tuberculosis_H37Rv ARAAVPSGASTGEHEAVELRDGGDRYGGKGVQKAVQAVLDEIGPAVIGLN
MUL_4631|M.ulcerans_Agy99 ARAAVPSGASTGEHEAVELRDGGDRYGGKGVKKAVEAVLDEIGPAVIGLN
MMAR_4462|M.marinum_M ARAAVPSGASTGEHEAVELRDGGDRYGGKGVKKAVEAVLDEIGPAVIGLN
MLBr_00255|M.leprae_Br4923 ARAAVPSGASTGEYEAVELRDGDGRYGGKGVKRAVDAVLDEIGPVVIGLN
MAV_1164|M.avium_104 ARAAVPSGASTGEHEAVELRDGGERYGGKGVQKAVQAVLDEIGPAVIGLN
Mflv_1954|M.gilvum_PYR-GCK ARAAVPSGASTGEHEAVELRDGGSRYLGKGVEKAVEAVLDEIAPAVIGLG
Mvan_4771|M.vanbaalenii_PYR-1 SRAAVPSGASTGEHEAVELRDGGSRYLGKGVEKAVEAVLDEIAPAVIGLG
MSMEG_5415|M.smegmatis_MC2_155 ARAAVPSGASTGEHEAVELRDGGSRYGGKGVEKAVEAVLDEIAPQVIGLS
MAB_1165|M.abscessus_ATCC_1997 ARAAVPSGASTGEHEAVELRDGDARYGGKGVTKAVNAVLDEIAPAIIGES
:************:********. ** **** :**:******.* :** .
Mb1051|M.bovis_AF2122/97 ADDQRLVDQALVDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFRY
Rv1023|M.tuberculosis_H37Rv ADDQRLVDQALVDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFRY
MUL_4631|M.ulcerans_Agy99 ADDQRLVDQALVDLDGTPDKSRLGGNSILGVSLAVAKAASESAELPLFRY
MMAR_4462|M.marinum_M ADDQRLVDQALVDLDGTPDKSRLGGNSILGVSLAVAKAASESAELPLFRY
MLBr_00255|M.leprae_Br4923 ANDQRLIDQELLDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFRY
MAV_1164|M.avium_104 ADDQRLVDQALVDLDGTPDKSRLGGNAILGVSLAVAKAAADSAELPLFRY
Mflv_1954|M.gilvum_PYR-GCK ADEQRLVDQALLDLDGTPDKSRLGANAILGVSLAVAKAAAQSAELPLFRY
Mvan_4771|M.vanbaalenii_PYR-1 ADEQRLVDQALVDLDGTPDKSRLGANAILGVSLAVAKAAAQSAELPLFRY
MSMEG_5415|M.smegmatis_MC2_155 ADDQRLVDQALLDLDGTPDKSRLGANAILGVSLAVSKAAAESAGLPLFRY
MAB_1165|M.abscessus_ATCC_1997 ADDQRLIDQALLDLDGTPDKSRLGANAILGVSLAVAKAAADSAGLALFRY
*::***:** *:************.*:********:***::** *.****
Mb1051|M.bovis_AF2122/97 VGGPNAHILPVPMMNILNGGAHADTAVDIQEFMVAPIGAPSFVEALRWGA
Rv1023|M.tuberculosis_H37Rv VGGPNAHILPVPMMNILNGGAHADTAVDIQEFMVAPIGAPSFVEALRWGA
MUL_4631|M.ulcerans_Agy99 IGGPNAHILPVPMMNILNGGAHADTGVDIQEFMVAPIGAPSFSEALRWGA
MMAR_4462|M.marinum_M IGGPNAHILPVPMMNILNGGAHADTGVDIQEFMVAPIGAPSFSEALRWGA
MLBr_00255|M.leprae_Br4923 IGGSNAHILPVPMMNILNGGAHADTAVDVQEFMVAPIGAPSFVEALRWGA
MAV_1164|M.avium_104 LGGPNAHILPVPMMNILNGGAHADTAVDIQEFMVAPIGAPSFAEALRWGA
Mflv_1954|M.gilvum_PYR-GCK LGGPNAHILPVPMMNIINGGAHADTGVDVQEFMIAPIGAPSFKEALRWGA
Mvan_4771|M.vanbaalenii_PYR-1 VGGPNAHILPVPMMNIINGGAHADTGVDVQEFMIAPIGAPSFKEALRWGA
MSMEG_5415|M.smegmatis_MC2_155 IGGPNAHILPVPMMNILNGGAHADTGVDVQEFMVAPIGAPSFKEALRWGA
MAB_1165|M.abscessus_ATCC_1997 LGGPNAHILPVPMMNILNGGAHADTGVDVQEFMVAPIGAPSFKESLRWGT
:**.************:********.**:****:******** *:****:
Mb1051|M.bovis_AF2122/97 EVYHALKSVLKKEGLSTGLGDEGGFAPDVAGTTAALDLISRAIESAGLRP
Rv1023|M.tuberculosis_H37Rv EVYHALKSVLKKEGLSTGLGDEGGFAPDVAGTTAALDLISRAIESAGLRP
MUL_4631|M.ulcerans_Agy99 EVYHALKAVLKKAGLSTGLGDEGGFAPDVASTTAALDLISQAIEAAGFKP
MMAR_4462|M.marinum_M EVYHALKAVLKKAGLSTGLGDEGGFAPDVASTTAALDLISQAIEAAGFKP
MLBr_00255|M.leprae_Br4923 EVYHALKSVLKKKGLSTGLGDEGGFAPEVAGTTAALDLVTLAIEAAGFKP
MAV_1164|M.avium_104 EVYHSLKSVLKKEGLSTGLGDEGGFAPDVAGTTAALDLIGRAIESAGFKL
Mflv_1954|M.gilvum_PYR-GCK EVYHSLKSVLKKQGLSTGLGDEGGFAPDLPGTKAALDLIGTAIEGAGLKI
Mvan_4771|M.vanbaalenii_PYR-1 EVYHSLKSVLKKQGLATGLGDEGGFAPDLPGTRAALDLIGTAIEAAGLKV
MSMEG_5415|M.smegmatis_MC2_155 EVYHSLKSVLKNQGLATGLGDEGGFAPDVAGTKAALDLISSAIEATGLKL
MAB_1165|M.abscessus_ATCC_1997 EVYHSLKSVLKKQGLSTGLGDEGGFAPDVAGTRAALDLISTAIEATGLKL
****:**:***: **:***********::..* *****: ***.:*::
Mb1051|M.bovis_AF2122/97 GADVALALDAAATEFFTDGTGYVFEGTTRTADQMTEFYAGLLGAYPLVSI
Rv1023|M.tuberculosis_H37Rv GADVALALDAAATEFFTDGTGYVFEGTTRTADQMTEFYAGLLGAYPLVSI
MUL_4631|M.ulcerans_Agy99 GVDVALALDAAANEFHADGS-YTFEGTPRTAAQMTEFYAGLLGSYPVVSI
MMAR_4462|M.marinum_M GVDVALALDAAANEFHADGS-YTFEGTPRTAAQMTEFYAGLLGSYPLVSI
MLBr_00255|M.leprae_Br4923 GADVALALDAAATEFYTDGIGYHFEGMTHTADQMTEFYADLLGSYPLVSI
MAV_1164|M.avium_104 GTDVALALDAAATEFYSDGTGYKFEGSTRTAEQMAEFYAGLLGAYPLVSI
Mflv_1954|M.gilvum_PYR-GCK GSDVALALDVAATEFYTDGTGYAFEKETRTAEQMAQFYEQLIGAYPLVSI
Mvan_4771|M.vanbaalenii_PYR-1 GSDVALALDVAATEFYTEGTGYAFEKETRTAEQMAAFYAQLLEAYPLVSI
MSMEG_5415|M.smegmatis_MC2_155 GSDVALALDVAATEFYTEGSGYAFEKETRTAEQMAEFYAGLLDSYPLVSI
MAB_1165|M.abscessus_ATCC_1997 GADVALALDVAATEFYSAADGYSFEKEKRTAEQMGAFYAELLDAYPLVSI
* *******.**.**.: . * ** :** ** ** *: :**:***
Mb1051|M.bovis_AF2122/97 EDPLSEDDWDGWAALTASIGDRVQIVGDDIFVTNPERLEEGIERGVANAL
Rv1023|M.tuberculosis_H37Rv EDPLSEDDWDGWAALTASIGDRVQIVGDDIFVTNPERLEEGIERGVANAL
MUL_4631|M.ulcerans_Agy99 EDPLYENDWDGWAALTAEIGDRVQIVGDDVFVTNPERLEEGIDRGVANAL
MMAR_4462|M.marinum_M EDPLYENDWDGWAALTAEIGDRVQIVGDDVFVTNPERLEEGIDRGVANAL
MLBr_00255|M.leprae_Br4923 EDPLSEDDWDGWAALTASIGEQVQIVGDDIFATNPERLEEGIGRGVANAL
MAV_1164|M.avium_104 EDPLSEDDWDGWAALTASIGDRVQLVGDDVFVTNPERLEEGIEKGVANAL
Mflv_1954|M.gilvum_PYR-GCK EDPLSEDDWDGWVALTSAIGDRVQLVGDDLFVTNPERLEDGIERGAGNAL
Mvan_4771|M.vanbaalenii_PYR-1 EDPLSEDDWDGWVALTAAIGDKVQLVGDDLFVTNPERLEDGIERGAANAL
MSMEG_5415|M.smegmatis_MC2_155 EDPLSEDDWDGWVSLTAAIGDRIQLVGDDLFVTNPERLEDGIQRGAANAL
MAB_1165|M.abscessus_ATCC_1997 EDPLSEDDWDGWVALTTAIGDRVQLVGDDLFVTNPERLEEGIERGAANAL
**** *:*****.:**: **:::*:****:*.*******:** :*..***
Mb1051|M.bovis_AF2122/97 LVKVNQIGTLTETLDAVTLAHHGGYRTMISHRSGETEDTMIADLAVAIGS
Rv1023|M.tuberculosis_H37Rv LVKVNQIGTLTETLDAVTLAHHGGYRTMISHRSGETEDTMIADLAVAIGS
MUL_4631|M.ulcerans_Agy99 LVKVNQIGTLTETLDAVALAHHSGYRTMISHRSGETEDTIIADLAVAVGS
MMAR_4462|M.marinum_M LVKVNQIGTLTETLDAVALAHHSGYRTMISHRSGETEDTIIADLAVAVGS
MLBr_00255|M.leprae_Br4923 LVKVNQIGTLTETLEAVALAHHSGYRTMISHRSGETEDTMIADLVVALGS
MAV_1164|M.avium_104 LVKVNQIGTLTETLDAVALAHHSGYRTMMSHRSGETEDTTIADLAVAVGS
Mflv_1954|M.gilvum_PYR-GCK LVKVNQIGTLTETLDAVSLAHHAGYKTMMSHRSGETEDTTIADLAVAVGS
Mvan_4771|M.vanbaalenii_PYR-1 LVKVNQIGTLTETLDAVSLAHNAGYKTMMSHRSGETEDTTIADLAVAVGS
MSMEG_5415|M.smegmatis_MC2_155 LVKVNQIGTLTETLDAVSLAHNSGYRTMMSHRSGETEDTTIADLAVAVGS
MAB_1165|M.abscessus_ATCC_1997 LVKVNQIGTLTETLDAVTLAHSSGYKTMMSHRSGETEDTTIADLAVAVGS
**************:**:*** .**:**:********** ****.**:**
Mb1051|M.bovis_AF2122/97 GQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFACETK-
Rv1023|M.tuberculosis_H37Rv GQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFACETK-
MUL_4631|M.ulcerans_Agy99 GQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFVADPK-
MMAR_4462|M.marinum_M GQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFVADPK-
MLBr_00255|M.leprae_Br4923 GQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFLRYVVETR-
MAV_1164|M.avium_104 GQIKTGAPARSERVAKYNQLLRIEEALGDAARYAGDLAFPRFALETR-
Mflv_1954|M.gilvum_PYR-GCK GQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFPRFGVESK-
Mvan_4771|M.vanbaalenii_PYR-1 GQIKTGAPARSERVAKYNQLLRIEEELGDAARYAGDLAFPRFSVETK-
MSMEG_5415|M.smegmatis_MC2_155 GQIKTGAPARSERVAKYNQLLRIEETLGDAARYAGDLAFPRLEAK---
MAB_1165|M.abscessus_ATCC_1997 GQIKTGAPARSERVAKYNQLLRIEETLGDAARFAGDLAFPRFSIEPAN
************************* ******:****** * .