For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASTADFKNGLVLVIDGQLWQIVEFQHVKPGKGPAFVRTKLKNVLSGKVVDKTYNAGVKVETATVDRRDTT YLYRDGSDFVFMDSQDYEQHPLPESLVGDAARFLLEGMPVQVAFHNGSPLYIELPVSVEMEVTHTEPGLQ GDRSSAGTKPATVETGAEIQVPLFINTGDKLKVDTRDGSYLGRVNA*
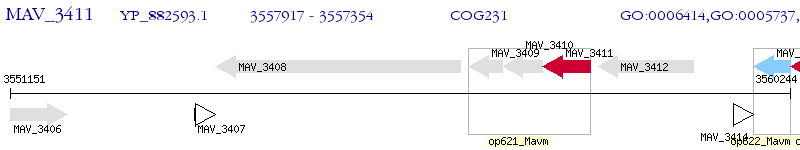
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3411 | efp | - | 100% (187) | elongation factor P |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2563c | efp | 1e-99 | 92.51% (187) | elongation factor P |
| M. gilvum PYR-GCK | Mflv_3752 | - | 3e-95 | 89.30% (187) | elongation factor P |
| M. tuberculosis H37Rv | Rv2534c | efp | 1e-99 | 92.51% (187) | elongation factor P |
| M. leprae Br4923 | MLBr_00522 | efp | 7e-98 | 90.86% (186) | elongation factor P |
| M. abscessus ATCC 19977 | MAB_2837c | - | 2e-93 | 90.32% (186) | elongation factor P |
| M. marinum M | MMAR_2181 | efp | 1e-99 | 93.58% (187) | elongation factor P Efp |
| M. smegmatis MC2 155 | MSMEG_3035 | efp | 7e-98 | 91.44% (187) | elongation factor P |
| M. thermoresistible (build 8) | TH_0105 | efp | 2e-97 | 90.81% (185) | PROBABLE ELONGATION FACTOR P EFP |
| M. ulcerans Agy99 | MUL_1765 | efp | 1e-99 | 93.58% (187) | elongation factor P |
| M. vanbaalenii PYR-1 | Mvan_2651 | - | 5e-96 | 90.37% (187) | elongation factor P |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2563c|M.bovis_AF2122/97 MATTADFKNGLVLVIDGQLWTITEFQHVKPGKGPAFVRTKLKNVLSGKVV
Rv2534c|M.tuberculosis_H37Rv MATTADFKNGLVLVIDGQLWTITEFQHVKPGKGPAFVRTKLKNVLSGKVV
MLBr_00522|M.leprae_Br4923 MATTADFKNGLVLVIDGQLWTIIGFQHVKPGKGPAFVRTKLKNVLSGKVV
MMAR_2181|M.marinum_M MASTADFKNGLVLQIDGQLWSIVEFQHVKPGKGPAFVRTKLKNVLSGKVV
MUL_1765|M.ulcerans_Agy99 MASTADFKNGLVLQIDGQLWSIVEFQHVKPGKGPAFVRTKLKNVLSGKVV
MAB_2837c|M.abscessus_ATCC_199 MASTADFKNGLVLNIDGQLWQITEFQHVKPGKGPAFVRTKLKNVLSGKVV
MAV_3411|M.avium_104 MASTADFKNGLVLVIDGQLWQIVEFQHVKPGKGPAFVRTKLKNVLSGKVV
Mflv_3752|M.gilvum_PYR-GCK MASTADFKNGLVLQIDGQLWQIVEFQHVKPGKGPAFVRTKLKNVVSGKVV
Mvan_2651|M.vanbaalenii_PYR-1 MASTADFKNGLVLQIDGQLWQIVEFQHVKPGKGPAFVRTKLKNVVSGKVV
MSMEG_3035|M.smegmatis_MC2_155 MASTADFKNGLVLQIDGQLWQIVEFQHVKPGKGPAFVRTKLKNVVSGKVV
TH_0105|M.thermoresistible__bu VATTADFKNGLVLVIDGQLWQIVEFLHVKPGKGPAFVRTKLKNVVSGKTV
:*:********** ****** * * ******************:***.*
Mb2563c|M.bovis_AF2122/97 DKTFNAGVKVDTATVDRRDTTYLYRDGSDFVFMDSQDYEQHPLPEALVGD
Rv2534c|M.tuberculosis_H37Rv DKTFNAGVKVDTATVDRRDTTYLYRDGSDFVFMDSQDYEQHPLPEALVGD
MLBr_00522|M.leprae_Br4923 DKTYNAGVKVDTATVDRRDTTYLYRDGANFVFMDSQDYEQHPLPESLVGD
MMAR_2181|M.marinum_M DKTYNAGVKVETATVDRRDTTYLYRDGSDFVFMDSQDYEQHPLPESLVGD
MUL_1765|M.ulcerans_Agy99 DKTYNAGVKVETATVDRRDTTYLYRDGSDFVFMDSQDYEQHPLPESLVGD
MAB_2837c|M.abscessus_ATCC_199 DKTFNAGVKVETATVDRRDTTYLYRDGDSFVFMDSQDYEQHHLSPELVGD
MAV_3411|M.avium_104 DKTYNAGVKVETATVDRRDTTYLYRDGSDFVFMDSQDYEQHPLPESLVGD
Mflv_3752|M.gilvum_PYR-GCK DKTYNAGVKVETATVDRRDATYLYRDGSDFVFMDSEDFEQHPLPEALVGR
Mvan_2651|M.vanbaalenii_PYR-1 DKTYNAGVKVETATVDRRDATYLYRDGSDFVFMDSEDYEQHPLPEALVGR
MSMEG_3035|M.smegmatis_MC2_155 DKTYNAGVKVETATVDRRDATYLYRDGSDFVFMDSEDFEQHPLPESLVGR
TH_0105|M.thermoresistible__bu DKTFNAGVKVETATVDRRDATYLYRDGSDFVFMDSENYEQHPLPESMVGD
***:******:********:******* .******:::*** *. :**
Mb2563c|M.bovis_AF2122/97 AARFLLEGMPVQVAFHNGVPLYIELPVTVELEVTHTEPGLQGDRSSAGTK
Rv2534c|M.tuberculosis_H37Rv AARFLLEGMPVQVAFHNGVPLYIELPVTVELEVTHTEPGLQGDRSSAGTK
MLBr_00522|M.leprae_Br4923 TARFLLEGMSVQVAFHNGVPLYVELPVTVELEVTHTEPGLQGDRSSAGTK
MMAR_2181|M.marinum_M AARFLLEGMPVQVAFHDGAPLYIELPVTVEIVVTHTEPGLQGDRSSAGTK
MUL_1765|M.ulcerans_Agy99 AARFLLEGMPVQVAFHDGAPLYIELPVTVEFVVTHTEPGLQGDRSSAGTK
MAB_2837c|M.abscessus_ATCC_199 AHRFLLEGMTVQVAFNEGAALYIELPVSVELEVTHTEPGLQGDRSSAGTK
MAV_3411|M.avium_104 AARFLLEGMPVQVAFHNGSPLYIELPVSVEMEVTHTEPGLQGDRSSAGTK
Mflv_3752|M.gilvum_PYR-GCK LAGFLLESMPVQIAFHDGVPLYLELPVTVELLVAHTEPGLQGDRSSAGTK
Mvan_2651|M.vanbaalenii_PYR-1 AADFLLESMPVQIAFHDGSPLYLELPVTVELVVASTEPGLQGDRSSAGTK
MSMEG_3035|M.smegmatis_MC2_155 LADFLLESMPVQIAFHDGTPLYLELPVSVELEVTHTEPGLQGDRSSAGTK
TH_0105|M.thermoresistible__bu AAKFLLESLPVQIAFHDGSPLYLELPVSVELEVTHTEPGLQGDRSSAGTK
****.:.**:**::* .**:****:**: *: ***************
Mb2563c|M.bovis_AF2122/97 PATLQTGAQINVPLFINTGDKLKVDSRDGSYLGRVNA
Rv2534c|M.tuberculosis_H37Rv PATLQTGAQINVPLFINTGDKLKVDSRDGSYLGRVNA
MLBr_00522|M.leprae_Br4923 PATLETGAQINVPLFINTGDKLKVDSRDGSYLGRVNV
MMAR_2181|M.marinum_M PATLETGAQINVPLFINTGDKLKVDSRDGGYLGRVNA
MUL_1765|M.ulcerans_Agy99 PATLETGAQINVPLFINTGDKLKVDSRDGGYLGRVNA
MAB_2837c|M.abscessus_ATCC_199 PATVETGAEIQVPLFINTGDRLKVDTRDASYLGRVNG
MAV_3411|M.avium_104 PATVETGAEIQVPLFINTGDKLKVDTRDGSYLGRVNA
Mflv_3752|M.gilvum_PYR-GCK PATMETGAEIQVPLFINTGDKLKVDSRDGSYLGRVNA
Mvan_2651|M.vanbaalenii_PYR-1 PATLETGAEIQVPLFINTGDKLKVDSRDGSYLGRVNA
MSMEG_3035|M.smegmatis_MC2_155 PATVETGAEIQVPLFINTGDRLKVDTRDGSYLGRVNA
TH_0105|M.thermoresistible__bu PATVETGAEIQVPLFINTGDKVKVDTRDGSYLGRVTN
***::***:*:*********::***:**..*****.