For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTRIEIFPDSQALVTAAAARLVDTIADAVAARGRALIVLTGGGNGIGLLKSLAGRPIDWSAVHLFWGDE RYVPEDDDERNEKQAREALLSHVDIPSSQVHPMPASDGEFGSDLAAAALAYEQLLAANAAPGQPVPNFDV HLLGMGPEGHINSLFPDTPAVLETTRMVVPVEDSPKPPPQRITLTLPAIARSREVWLMVSGAGKADAVAA AINGAAPVSVPAAGAVGLDTTLWLLDREAAAKLPPGTAVQG
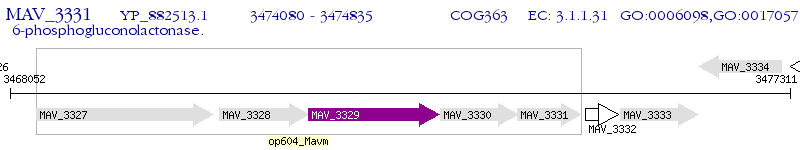
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3331 | pgl | - | 100% (251) | 6-phosphogluconolactonase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1480c | devB | 1e-108 | 79.67% (246) | 6-phosphogluconolactonase |
| M. gilvum PYR-GCK | Mflv_3696 | - | 2e-87 | 66.53% (245) | 6-phosphogluconolactonase |
| M. tuberculosis H37Rv | Rv1445c | devB | 1e-108 | 79.67% (246) | 6-phosphogluconolactonase |
| M. leprae Br4923 | MLBr_00579 | pgl | 1e-105 | 76.83% (246) | putative 6-phosphogluconolactonase |
| M. abscessus ATCC 19977 | MAB_2763 | - | 3e-87 | 67.08% (243) | 6-phosphogluconolactonase |
| M. marinum M | MMAR_2250 | devB | 1e-105 | 76.83% (246) | 6-phosphogluconolactonase DevB |
| M. smegmatis MC2 155 | MSMEG_3099 | pgl | 4e-88 | 67.49% (243) | 6-phosphogluconolactonase |
| M. thermoresistible (build 8) | TH_0335 | pgl | 7e-92 | 68.57% (245) | 6-phosphogluconolactonase |
| M. ulcerans Agy99 | MUL_1840 | devB | 1e-105 | 76.83% (246) | 6-phosphogluconolactonase DevB |
| M. vanbaalenii PYR-1 | Mvan_2716 | - | 2e-87 | 67.76% (245) | 6-phosphogluconolactonase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3696|M.gilvum_PYR-GCK -MTMNIETYPDTEALVAATGDRLVEAIKAAIAARDVAAIALTGGGTGIGL
Mvan_2716|M.vanbaalenii_PYR-1 -MTREIETYSGTAALVAAAGDRLVASIKNAVEGRSSAHIVLTGGGTGIGL
MSMEG_3099|M.smegmatis_MC2_155 MSDTVIERHADTAALVAAAGDRLVDAISSAIGERGQATIVLTGGGTGIGL
TH_0335|M.thermoresistible__bu -MSTFVETHPDTATLVTAAGDRLVEAINAATSARGEAVIVLTGGGSGIAL
Mb1480c|M.bovis_AF2122/97 -MSSSIEIFPDSDILVAAAGKRLVGAIGAAVAARGQALIVLTGGGNGIAL
Rv1445c|M.tuberculosis_H37Rv -MSSSIEIFPDSDILVAAAGKRLVGAIGAAVAARGQALIVLTGGGNGIAL
MMAR_2250|M.marinum_M -MSLDVEIFPDSDMLIGAAGQRLIQTIGSAVAERGQALIVLTGGGNGIGL
MUL_1840|M.ulcerans_Agy99 -MSLDVEIFPDSDMLIGAAGQRLIQTIGSAVAERGQALIVLTGGGNGIGL
MAV_3331|M.avium_104 -MSTRIEIFPDSQALVTAAAARLVDTIADAVAARGRALIVLTGGGNGIGL
MLBr_00579|M.leprae_Br4923 -MSASVEIFSDSKTMVGAAGKRLASTIQSAVAARERALIVLTGGSSGIGL
MAB_2763|M.abscessus_ATCC_1997 MSETIIEKYADTDALVTAAGDRLASAITGALAERGKAMIVLTGGGTGIAL
:* ...: :: *:. ** :* * * * *.****..**.*
Mflv_3696|M.gilvum_PYR-GCK LRRVGERGDEIDWSKVHIYWGDERFVPADDDERNEKQAREALLDHVGIPE
Mvan_2716|M.vanbaalenii_PYR-1 LGRVAERSGEIDWSKVHIYWGDERFVAHDDDERNEKQAREALLDHVDIPA
MSMEG_3099|M.smegmatis_MC2_155 LKRVRERSGEIDWSKVHIYWGDERFVPQDDDERNDKQAREALLDHIGIPP
TH_0335|M.thermoresistible__bu LKRVGERAGEIDWSKVRLYWGDERYVPAADDERNEKQAREALLDHIDIPA
Mb1480c|M.bovis_AF2122/97 LRYLSAQAQQIEWSKVHLFWGDERYVPEDDDERNLKQARRALLNHVDIPS
Rv1445c|M.tuberculosis_H37Rv LRYLSAQAQQIEWSKVHLFWGDERYVPEDDDERNLKQARRALLNHVDIPS
MMAR_2250|M.marinum_M LRYLGSQGPQIDWSKVHLFWGDDRYVPEDDDERNDKQAREALLDHVDIPA
MUL_1840|M.ulcerans_Agy99 LRYLGSQGPQIDWSKVHLFWGDDRYVPEDDDERNDKQAREALLDHVDIPA
MAV_3331|M.avium_104 LKSLAGR--PIDWSAVHLFWGDERYVPEDDDERNEKQAREALLSHVDIPS
MLBr_00579|M.leprae_Br4923 LRDLATRGQQIDWSRVHLFWGDERYVPKDDDERNEKQARVALLDHIDIPP
MAB_2763|M.abscessus_ATCC_1997 LKHLRDVASGLDWTNVHVFWGDDRYVPKTDPERNAWQAWEALLEHVNFPL
* : ::*: *:::***:*:*. * *** ** ***.*:.:*
Mflv_3696|M.gilvum_PYR-GCK ANVHVMAPSDGEFGDAIDAAAEAYAQVLAEAG--PDPTPAFDVHLLGMGP
Mvan_2716|M.vanbaalenii_PYR-1 ANVHAMAASDGEFGDALDAAAEAYAQLLTEAGG-AEPTPAFDVHLLGMGG
MSMEG_3099|M.smegmatis_MC2_155 VNVHAMAASDGEFGDDLEAAAAGYAQLL-SANF-DSSVPGFDVHLLGMGG
TH_0335|M.thermoresistible__bu ANVHPMAASDGEFGDDIAAAAAAYEQVL---------PAEFDVHLLGMGG
Mb1480c|M.bovis_AF2122/97 NQVHPMAASDGDFGGDLDAAALAYEQVLAASAAPGDPAPNFDVHLLGMGP
Rv1445c|M.tuberculosis_H37Rv NQVHPMAASDGDFGGDLDAAALAYEQVLAASAAPGDPAPNFDVHLLGMGP
MMAR_2250|M.marinum_M SQVHPMAASDGEFGADLDAAALAYEQVLAASAVPGNRAPDFDVHLLGMGP
MUL_1840|M.ulcerans_Agy99 SQVHPMAASDGEFGADLDAAALAYEQVLAASAVPGNRAPDFDVHLLGMGP
MAV_3331|M.avium_104 SQVHPMPASDGEFGSDLAAAALAYEQLLAANAAPGQPVPNFDVHLLGMGP
MLBr_00579|M.leprae_Br4923 SQVHPMPAGDGEFGNDLEAAALAYEQLLAAYAAPGYPTPNFDVHLMGMGP
MAB_2763|M.abscessus_ATCC_1997 RNMHAMPNSESEYGTDLDAAALAYEQLLAANAEPGQDCPAFDVHLLGMGG
::* *. .:.::* : *** .* *:* . *****:***
Mflv_3696|M.gilvum_PYR-GCK EGHVNSLFPDTDAVKESKRFVVGVTDCPKPPPRRITLTLPAVQRSREVWL
Mvan_2716|M.vanbaalenii_PYR-1 EGHVNSLFPDTAAVRETERLVVGVADSPKPPPRRITLTLPAITRSREVWL
MSMEG_3099|M.smegmatis_MC2_155 EGHVNSLFPDTDAVRETERLVVGVSDSPKPPPRRITLTLPAVQNSREVWL
TH_0335|M.thermoresistible__bu EGHINSLFPDTPAVRETERLVVPVTDSPKPPPRRISLTLPAVRRARQVWL
Mb1480c|M.bovis_AF2122/97 EGHINSLFPHSPAVLESTRMVVAVDDSPKPPPRRITLTLPAIQRSREVWL
Rv1445c|M.tuberculosis_H37Rv EGHINSLFPHSPAVLESTRMVVAVDDSPKPPPRRITLTLPAIQRSREVWL
MMAR_2250|M.marinum_M EGHINSLFPGTQAVRETTRLVVPVGDSPKPPPQRITLTLPAIQRSREVWL
MUL_1840|M.ulcerans_Agy99 EGHINSLFPGTQAVRETTRLVVPVGDSPKPPPQRITLTLPAIQRSREVWL
MAV_3331|M.avium_104 EGHINSLFPDTPAVLETTRMVVPVEDSPKPPPQRITLTLPAIARSREVWL
MLBr_00579|M.leprae_Br4923 EGHINSLFPNTVAVRETSRMVVGVRNSPKPPPERITLTLNAIQRSREVWL
MAB_2763|M.abscessus_ATCC_1997 EGHINSLFPHTDAVKETQRLVVAVPDSPKPPPQRITLTLPAIQRSREVWL
***:***** : ** *: *:** * :.*****.**:*** *: .:*:***
Mflv_3696|M.gilvum_PYR-GCK VVSGEGKADAVAAAIGGAQPVDIPAAGAVGTEATVWLLDEAAASKL----
Mvan_2716|M.vanbaalenii_PYR-1 VVSGAAKADAVAAAIGGADPVDIPAAGAVGTEATVWLLDEDAASKL----
MSMEG_3099|M.smegmatis_MC2_155 VVSGEAKADAVAAAVGGADPVDIPAAGAVGRERTVWLVDEAAAAKL----
TH_0335|M.thermoresistible__bu VVSGAAKADAVAAAVNGADPVDIPAAGAVGREATVWLLDEEAASRLDR--
Mb1480c|M.bovis_AF2122/97 LVSGPGKADAVAAAIGGADPVSVPAAGAVGRQNTLWLLDRDAAAKLPS--
Rv1445c|M.tuberculosis_H37Rv LVSGPGKADAVAAAIGGADPVSVPAAGAVGRQNTLWLLDRDAAAKLPS--
MMAR_2250|M.marinum_M MVSGGTKAEAVAAAIGGADPVLLPAAGAIGRESTRWLLDRDAAARLPG--
MUL_1840|M.ulcerans_Agy99 MVSGGTKAEAVAAAIGGADPVLLPAAGAIGRESTRWLLDRDAAARLPG--
MAV_3331|M.avium_104 MVSGAGKADAVAAAINGAAPVSVPAAGAVGLDTTLWLLDREAAAKLPPGT
MLBr_00579|M.leprae_Br4923 MVSGTAKADAVAAAMGGAPSASIPAAGAVGLETTLWLLDEEAAAKIPG--
MAB_2763|M.abscessus_ATCC_1997 VVSGEAKADAVAAAVGGADPVDVPAAGAKGIERTVWLLDEAAASQLG---
:*** **:*****:.** .. :***** * : * **:*. **:::
Mflv_3696|M.gilvum_PYR-GCK ----
Mvan_2716|M.vanbaalenii_PYR-1 ----
MSMEG_3099|M.smegmatis_MC2_155 ----
TH_0335|M.thermoresistible__bu ----
Mb1480c|M.bovis_AF2122/97 ----
Rv1445c|M.tuberculosis_H37Rv ----
MMAR_2250|M.marinum_M ----
MUL_1840|M.ulcerans_Agy99 ----
MAV_3331|M.avium_104 AVQG
MLBr_00579|M.leprae_Br4923 ----
MAB_2763|M.abscessus_ATCC_1997 ----