For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDHDTRFAYLDLLRRDLTRYGSDELVPVGLYRLGRPLFSTRNLMLVRKRPFNKQARDLGLDWPADALTMI GMKRLTSLQHCVETVLEEDVPGDLVECGVWRGGASILMRAVLAAYGDEKRCVWLCDSFAGVPPPDTVNYK ADKGIRLHRHARILGVPLENVKANFERYGLLDDQVRFVPGWFKDTLKDAPIDRISVLRLDGDLYESTIQA LDALYPRLSPGGFCIVDDYHAIKACAQAVTDYRTQHGVTAEIVEIDGTGVLWRKP*
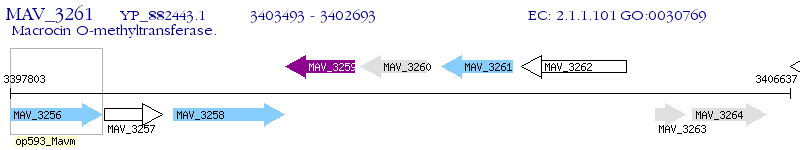
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3261 | - | - | 100% (266) | methyltransferase MtfC |
| M. avium 104 | MAV_3266 | - | e-132 | 85.06% (261) | MtfB protein |
| M. avium 104 | MAV_3260 | - | 1e-08 | 29.66% (145) | MtfD protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4108c | - | 1e-122 | 79.25% (265) | methyltransferase MtfB |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0388 | tylF | 1e-111 | 80.33% (239) | macrocin-O-methyltransferase |
| M. thermoresistible (build 8) | TH_1991 | tylF | 1e-123 | 79.25% (265) | Macrocin-O-methyltransferase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4108c|M.abscessus_ATCC_199 MTVTDYDTRSAYLDLLRQDLTRYGVDELVPVGWNWLHRPFFKFGDYVLVR
TH_1991|M.thermoresistible__bu --VTDRDSRSAYLDLLRRDLTRYGSDELVPVGWNWLHRPVFKIGNLMLVR
MSMEG_0388|M.smegmatis_MC2_155 ----------------------------MPVGWDWFHRPLFKIGNLMLVR
MAV_3261|M.avium_104 --MTDHDTRFAYLDLLRRDLTRYGSDELVPVGLYRLGRPLFSTRNLMLVR
:*** : **.*. : :***
MAB_4108c|M.abscessus_ATCC_199 KRAFDAHKRDLGLDWPADALTMIGMQRLTSLQQCVETVLADDVPGDLVEC
TH_1991|M.thermoresistible__bu RRPFDKHKRDLGLDWPVEALTMIGMKRLTSLQECVETVLRDDVPGDLVEC
MSMEG_0388|M.smegmatis_MC2_155 RRPFDPRKRDLGLDWPADALTMIGMTRLTSLQQCVETVITEDVPGDLVEC
MAV_3261|M.avium_104 KRPFNKQARDLGLDWPADALTMIGMKRLTSLQHCVETVLEEDVPGDLVEC
:*.*: : ********.:******* ******.*****: :*********
MAB_4108c|M.abscessus_ATCC_199 GVWRGGASILMRAVLAAYGDTTRSVWLCDSFEGVPPPDTANYKADKGDRL
TH_1991|M.thermoresistible__bu GVWRGGASILMRAVLAAYGDEERCVWLADSFAGVPPPDAEKYRADQGDRL
MSMEG_0388|M.smegmatis_MC2_155 GVWRGGASILMRAVLSAYGDESRRVWLCDSFAGVPAPDAAKYKADKGDRL
MAV_3261|M.avium_104 GVWRGGASILMRAVLAAYGDEKRCVWLCDSFAGVPPPDTVNYKADKGIRL
***************:**** * ***.*** ***.**: :*:**:* **
MAB_4108c|M.abscessus_ATCC_199 HLAAPILAVTEKNVRANFERYGLLDQQVRFVPGWFKDTLHDAPIEQIAVL
TH_1991|M.thermoresistible__bu HLAAPILAVSEEEVRANFERYNLLDDQVRFLPGWFKDTLHKAPIEQIAVL
MSMEG_0388|M.smegmatis_MC2_155 HLAAPILAVSEQDVRANFQRYGLLDDQVRFVPGWFKDTLHDAPIERISIL
MAV_3261|M.avium_104 HRHARILGVPLENVKANFERYGLLDDQVRFVPGWFKDTLKDAPIDRISVL
* * **.*. ::*:***:**.***:****:********:.***::*::*
MAB_4108c|M.abscessus_ATCC_199 RLDGDLYESTIQALDGLYARLSPGGFCIIDDYHALDSCKQAVTDYRAKHG
TH_1991|M.thermoresistible__bu RLDGDLYESTIQALDGLYARLSPGGFCIIDDYHAIDGCRQAVTDFRATHG
MSMEG_0388|M.smegmatis_MC2_155 RLDGDLYESTIQALDGLYERLSPGGFCIVDDYHAIDGCRQAVTDYRAKHG
MAV_3261|M.avium_104 RLDGDLYESTIQALDALYPRLSPGGFCIVDDYHAIKACAQAVTDYRTQHG
***************.** *********:*****:..* *****:*: **
MAB_4108c|M.abscessus_ATCC_199 ITAEIMEIDGTGVFWRKE
TH_1991|M.thermoresistible__bu ITEDIVEIDGTGVLWRKD
MSMEG_0388|M.smegmatis_MC2_155 ISAEIVEIDGTGVLWRKE
MAV_3261|M.avium_104 VTAEIVEIDGTGVLWRKP
:: :*:*******:***