For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGQRATPQPTLDDLPLRDDLRGKSPYGAPQLAVPVRLNTNENPHPPSRALVDDVVRSVARAAADLHRYPD RDAVQLRSDLARYLTAQTGVQLGVENLWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIISDGTRT EWLQAARADDFSLDVDAAVAAVTERTPDVVFVASPNNPSGQSVSLSGLRRLLDAAPGIVIVDEAYGEFSS QPSAVQLVGEYPTKLVVTRTMSKAFAFAGGRLGYLIATPAVIEAMLLVRLPYHLSSVTQAAARAALRHAD DTLGSVAALIAERERVSTALTGMGFRVIPSDANFVLFGEFTDAPASWQRYLDAGVLIRDVGIPGYLRATT GLAEENDAFLRASAQLAATELAPVNVGAIASAAEPRAAGRDHVLGAP*
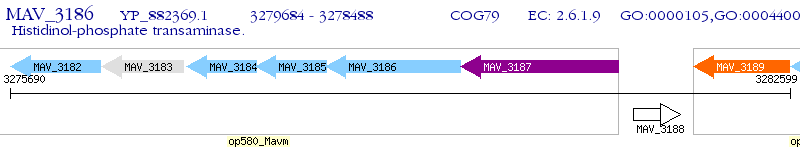
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3186 | hisC | - | 100% (398) | histidinol-phosphate aminotransferase |
| M. avium 104 | MAV_0250 | hisC | 2e-38 | 33.79% (364) | putative aminotransferase |
| M. avium 104 | MAV_4421 | - | 2e-29 | 29.79% (339) | histidinol-phosphate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1626 | hisC1 | 0.0 | 86.85% (365) | histidinol-phosphate aminotransferase |
| M. gilvum PYR-GCK | Mflv_3612 | - | 1e-167 | 80.17% (363) | histidinol-phosphate aminotransferase |
| M. tuberculosis H37Rv | Rv1600 | hisC1 | 0.0 | 86.85% (365) | histidinol-phosphate aminotransferase |
| M. leprae Br4923 | MLBr_01258 | hisC | 0.0 | 84.74% (367) | histidinol-phosphate aminotransferase |
| M. abscessus ATCC 19977 | MAB_2669c | - | 1e-160 | 76.18% (361) | histidinol-phosphate aminotransferase |
| M. marinum M | MMAR_2396 | hisC1 | 0.0 | 82.67% (375) | histidinol-phosphate aminotransferase HisC1 |
| M. smegmatis MC2 155 | MSMEG_3206 | hisC | 1e-172 | 83.56% (365) | histidinol-phosphate aminotransferase |
| M. thermoresistible (build 8) | TH_3588 | - | 1e-166 | 80.99% (363) | PUTATIVE histidinol-phosphate aminotransferase |
| M. ulcerans Agy99 | MUL_1573 | hisC1 | 1e-179 | 81.60% (375) | histidinol-phosphate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_2805 | - | 1e-168 | 81.27% (363) | histidinol-phosphate aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3612|M.gilvum_PYR-GCK MTVFEAGGPGFGVTLDDLPLRDDLRGKSPYGAPQLVVPVRLNTNENPHPP
Mvan_2805|M.vanbaalenii_PYR-1 ----MTGVPGSSITLDDLPLRDDLRGKSPYGAPQLSVPVRLNTNENPHPP
MMAR_2396|M.marinum_M ---MTSA-PRPRPTLDDLPLREDLRGKSPYGAPQLAVPVRLNTNENPHPP
MUL_1573|M.ulcerans_Agy99 ---MTSA-PRPRPTLDDLPLREDLRGKSPYGASQLAVPVRLSTNENPHPP
Mb1626|M.bovis_AF2122/97 ---MTRS-GHP-VTLDDLPLRADLRGKAPYGAPQLAVPVRLNTNENPHPP
Rv1600|M.tuberculosis_H37Rv ---MTRS-GHP-VTLDDLPLRADLRGKAPYGAPQLAVPVRLNTNENPHPP
MLBr_01258|M.leprae_Br4923 ---MN----VPEPTLDDLPLRDNLRGKSPYGAMQLLVPVLLNTNENPHPP
MAV_3186|M.avium_104 ---MTGQRATPQPTLDDLPLRDDLRGKSPYGAPQLAVPVRLNTNENPHPP
MSMEG_3206|M.smegmatis_MC2_155 -----MS--ADKVTLADLPLRDNLRGKSPYGAPQLQVPVRLNTNENPHPP
TH_3588|M.thermoresistible__bu ---MSIP--GRDVTLADLPLRENLRGKSPYGAPQLEVQVRLNTNENPHPP
MAB_2669c|M.abscessus_ATCC_199 ---MSVP-IGGSARLEDLPLRDSLRGKSPYGAPQLTVPVRLNTNENPHEP
* ***** .****:**** ** * * *.****** *
Mflv_3612|M.gilvum_PYR-GCK TQALIDDVARSVREVAGELHRYPDRDAVALRTDLAAYLTAQTGNPVGVEN
Mvan_2805|M.vanbaalenii_PYR-1 TRALIDDVAQSVREVAGELHRYPDRDAVALRTDLAAYLSAQTGTAVGVEN
MMAR_2396|M.marinum_M TQALVDDVVRSVGEAAVDLHRYPDRDAVALRTDLANYLTAQTGTRIGFEN
MUL_1573|M.ulcerans_Agy99 TQALVDDVVRSVGEAAVDLHRYPDRDAVALRTDLANYLTAQTGTRIGFEN
Mb1626|M.bovis_AF2122/97 TRALVDDVVRSVREAAIDLHRYPDRDAVALRADLAGYLTAQTGIQLGVEN
Rv1600|M.tuberculosis_H37Rv TRALVDDVVRSVREAAIDLHRYPDRDAVALRADLAGYLTAQTGIQLGVEN
MLBr_01258|M.leprae_Br4923 TKALVDDVVRSVQKVAVDLHRYPDRDAVALRQDLASYLTAQTGIRLGVEN
MAV_3186|M.avium_104 SRALVDDVVRSVARAAADLHRYPDRDAVQLRSDLARYLTAQTGVQLGVEN
MSMEG_3206|M.smegmatis_MC2_155 SKALVDDVAASVREAAAELHRYPDRDAVALRTDLAAYLTAATGVRLGVEN
TH_3588|M.thermoresistible__bu SQALIDDLTAAVRAEAAQLHRYPDRDAVALRTDLAEYLSSQTGVQLGMEN
MAB_2669c|M.abscessus_ATCC_199 SAGLVDDVAASIREVAGELHRYPDRDAVALRTDLATYLTEQTGVPVVAEN
: .*:**:. :: * :********** ** *** **: ** : **
Mflv_3612|M.gilvum_PYR-GCK VWAANGSNEILQQLLQAFGGPGRSALGFVPSYSMHPIIADGTQTRWLAAN
Mvan_2805|M.vanbaalenii_PYR-1 VWAANGSNEILQQLLQAFGGPGRSALGFVPSYSMHPIISDGTQTRWLVAN
MMAR_2396|M.marinum_M VWAANGSNEILQQLLQAFGGPGRTAIGFVPSYSMHPIISDGTHTEWVETA
MUL_1573|M.ulcerans_Agy99 VWAANGSNEILQQLLQAFGGPGRTAIGFVPSYSMHPIISDGTHTEWVETA
Mb1626|M.bovis_AF2122/97 IWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIISDGTHTEWIEAS
Rv1600|M.tuberculosis_H37Rv IWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIISDGTHTEWIEAS
MLBr_01258|M.leprae_Br4923 IWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIIADGTHTEWLETV
MAV_3186|M.avium_104 LWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIISDGTRTEWLQAA
MSMEG_3206|M.smegmatis_MC2_155 LWAANGSNEILQQLLQAFGGPGRTAIGFVPSYSMHPIISDGTQTEWLQAS
TH_3588|M.thermoresistible__bu LWAANGSNEILQQLLQAFGGPGRRAMGFVPSYSMHPIISDGTQTEWVQAA
MAB_2669c|M.abscessus_ATCC_199 VWAANGSNEVLQQLLQAFGGPGRRALGFTPSYSMHPIISDGTQTEWLQAQ
:********:************* *:**.*********:***:*.*: :
Mflv_3612|M.gilvum_PYR-GCK RADDFSLDAAAAATAIKEHAPDVVFVTSPNNPTGQSVSLDDLRVLLDALS
Mvan_2805|M.vanbaalenii_PYR-1 RGDDFGLDAAVAATAIKEHTPDVVFVTSPNNPSGQSVSLDDLRLLLDALC
MMAR_2396|M.marinum_M RADGFGLDIDAAIAVVSDRRPDVVFITSPNNPTGQSVSLTELRRLLDVVP
MUL_1573|M.ulcerans_Agy99 RADGFGLDIDAAIAVVSDRRPDVVFITSPNNPTGQSVSLTELRRLLDVVP
Mb1626|M.bovis_AF2122/97 RANDFGLDVDVAVAAVVDRKPDVVFIASPNNPSGQSVSLPDLCKLLDVAP
Rv1600|M.tuberculosis_H37Rv RANDFGLDVDVAVAAVVDRKPDVVFIASPNNPSGQSVSLPDLCKLLDVAP
MLBr_01258|M.leprae_Br4923 RADDFSLDVEAAVTAVADRKPDVVFIASPNNPSGQSISLADLRRLLDVVP
MAV_3186|M.avium_104 RADDFSLDVDAAVAAVTERTPDVVFVASPNNPSGQSVSLSGLRRLLDAAP
MSMEG_3206|M.smegmatis_MC2_155 RAEDFGLDIDVAVSAVTERKPDVVFVTSPNNPSGQSVPLDDLRRVLDAMQ
TH_3588|M.thermoresistible__bu RAADFGLDVDVAVRAITEQRPDVVFVASPNNPSGQSVTPDELRALLDAMP
MAB_2669c|M.abscessus_ATCC_199 RSEDFSLDVGSAVAAVRERKPDVVFVTSPNNPTGQSIPLDALRRILDVAP
*. .*.** * .: :: *****::*****:***:. * :**.
Mflv_3612|M.gilvum_PYR-GCK SQSGGIMIVDEAYGEFSSQPSAIALLAQYPTRLVVSRTMSKAFAFAGGRL
Mvan_2805|M.vanbaalenii_PYR-1 LQDGGVMIVDEAYGEFSSQPSAIGLIESYPGKLVVTRTMSKAFAFAGGRV
MMAR_2396|M.marinum_M ----GILIVDEAYGEFSSQPSAVGLIEEYPTRLVVTRTMSKAFAFAGGRL
MUL_1573|M.ulcerans_Agy99 ----GILIVDEAYGEFSSQPSAVGLIEEYPTRVVVTRTMSKAFAFAGGRL
Mb1626|M.bovis_AF2122/97 ----GIAIVDEAYGEFSSQPSAVSLVEEYPSKLVVTRTMSKAFAFAGGRL
Rv1600|M.tuberculosis_H37Rv ----GIAIVDEAYGEFSSQPSAVSLVEEYPSKLVVTRTMSKAFAFAGGRL
MLBr_01258|M.leprae_Br4923 ----GILIVDEAYGEFSSRPSAVALVGEYPTKIVVTRTTSKAFAFAGGRL
MAV_3186|M.avium_104 ----GIVIVDEAYGEFSSQPSAVQLVGEYPTKLVVTRTMSKAFAFAGGRL
MSMEG_3206|M.smegmatis_MC2_155 G---GILIVDEAYGEFSSQPSAVALLDDYPAKLVVSRTMSKAFAFAGGRL
TH_3588|M.thermoresistible__bu R---GILIVDEAYGEFSSQPSAVGLIADYPTKLVVSRTMSKAFAFAGGRL
MAB_2669c|M.abscessus_ATCC_199 ----GVVILDEAYGEFSAAPSGITLVDEYPAKLIVSRTMSKAFAFAGGRL
*: *:********: **.: *: .** :::*:** **********:
Mflv_3612|M.gilvum_PYR-GCK GYLVAAPAVIDAMLLVRLPYHLSSVTQAAARAALRHAEETLSSVATLVAE
Mvan_2805|M.vanbaalenii_PYR-1 GYLVAAPAVIDAMLLVRLPYHLSSVTQAAARAALRHADDTLGSVATLIAE
MMAR_2396|M.marinum_M GYLVATPALIDALLLVRLPYHLSSVTQVAARAALRHAQDTLGSVATLIAE
MUL_1573|M.ulcerans_Agy99 GYLVATPALIDALLLVRLPYHLSSVTQVAARAALRHAQDTLGSVATLIAE
Mb1626|M.bovis_AF2122/97 GYLIATPAVIDAMLLVRLPYHLSSVTQAAARAALRHSDDTLSSVAALIAE
Rv1600|M.tuberculosis_H37Rv GYLIATPAVIDAMLLVRLPYHLSSVTQAAARAALRHSDDTLSSVAALIAE
MLBr_01258|M.leprae_Br4923 GYLIATPALVEAMLLVRLPYHLSSVTQAAARAALRHADDTLGSVAALIAE
MAV_3186|M.avium_104 GYLIATPAVIEAMLLVRLPYHLSSVTQAAARAALRHADDTLGSVAALIAE
MSMEG_3206|M.smegmatis_MC2_155 GYLAAAPAVIDALLLVRLPYHLSVLTQAAARAALRHADDTLGSVKALIAE
TH_3588|M.thermoresistible__bu GYLAAAPALIDALLLVRLPYHLSSLTQAAARAALRHADDTLAGVATLIAE
MAB_2669c|M.abscessus_ATCC_199 GYLVAAPAVIDALLLVRLPYHLSVLTQAAARAAVKHRAETLGSVATLVAE
*** *:**:::*:********** :**.*****::* :**..* :*:**
Mflv_3612|M.gilvum_PYR-GCK RERVSTALTELGFRVIPSDSNFVLFGEFADAPATWQRYLDEGVLIRDVGI
Mvan_2805|M.vanbaalenii_PYR-1 RERVSQALTGMGFRVIPSDANFILFGEFADAPATWQRYLDEGVLIRDVGI
MMAR_2396|M.marinum_M RERVSKKLAGMGFRVIPSDANFVLFGEFADAPAAWQRYLDQGVLIRDVGI
MUL_1573|M.ulcerans_Agy99 RERVSKKLASMGFRVIPSDANFVLFGEFADAPAAWQRYLDQGVLIRDVGI
Mb1626|M.bovis_AF2122/97 RERVTTSLNDMGFRVIPSDANFVLFGEFADAPAAWRRYLEAGILIRDVGI
Rv1600|M.tuberculosis_H37Rv RERVTTSLNDMGFRVIPSDANFVLFGEFADAPAAWRRYLEAGILIRDVGI
MLBr_01258|M.leprae_Br4923 RERVTKSLVHMGFRVIPSDANFVLFGHFSDAAGAWQHYLDTGVLIRDVGI
MAV_3186|M.avium_104 RERVSTALTGMGFRVIPSDANFVLFGEFTDAPASWQRYLDAGVLIRDVGI
MSMEG_3206|M.smegmatis_MC2_155 RERVSAELTRMGYRVIPSDANFVLFGAFTDAPATWQRYLDAGVLIRDVGI
TH_3588|M.thermoresistible__bu RERVSEELRRMGFRVIPSDANFVLFGEFADAPATWQRYLDDGVLIRDVGI
MAB_2669c|M.abscessus_ATCC_199 RIRVSEALSASGFRVIPSDANFILFGRFADAAASWRRYLDAGVLIRDVGI
* **: * *:******:**:*** *:**..:*::**: *:*******
Mflv_3612|M.gilvum_PYR-GCK PGYLRTTIGLADENDALLAASARIGAP-----------------------
Mvan_2805|M.vanbaalenii_PYR-1 PGYLRTTIGLAEENDALLAASARIGAP-----------------------
MMAR_2396|M.marinum_M PGYLRATTGLADENDAFLRASARIAATDLAPAAASPVGAP----------
MUL_1573|M.ulcerans_Agy99 PGYLRATTGLADENDAFLRASARIAATDLAPAAASPVGAP----------
Mb1626|M.bovis_AF2122/97 PGYLRATTGLAEENDAFLRASARIA-TDLVPVTRSPVGAP----------
Rv1600|M.tuberculosis_H37Rv PGYLRATTGLAEENDAFLRASARIA-TDLVPVTRSPVGAP----------
MLBr_01258|M.leprae_Br4923 PGYLRATTGLAEENDAFLKASSEIAATELAPATT--LGAS----------
MAV_3186|M.avium_104 PGYLRATTGLAEENDAFLRASAQLAATELAPVNVGAIASAAEPRAAGRDH
MSMEG_3206|M.smegmatis_MC2_155 PGHLRVTIGLAAENDAFLAAGAELAGTELQPS-TSPVGAQ----------
TH_3588|M.thermoresistible__bu PGHLRTTIGLAHENDALLATSARLAATELKAAPSSPIGAP----------
MAB_2669c|M.abscessus_ATCC_199 PGYLRVTVGLASENDVFLKVSDSLAATDLEVEAS----------------
**:**.* *** ***.:* .. :. .
Mflv_3612|M.gilvum_PYR-GCK -----
Mvan_2805|M.vanbaalenii_PYR-1 -----
MMAR_2396|M.marinum_M -----
MUL_1573|M.ulcerans_Agy99 -----
Mb1626|M.bovis_AF2122/97 -----
Rv1600|M.tuberculosis_H37Rv -----
MLBr_01258|M.leprae_Br4923 -----
MAV_3186|M.avium_104 VLGAP
MSMEG_3206|M.smegmatis_MC2_155 -----
TH_3588|M.thermoresistible__bu -----
MAB_2669c|M.abscessus_ATCC_199 -----