For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPELPDVEGFRRQLADALPGRRVRRVKVHDPGILRNTTATTLARRLTGRRFAGPRRHGKWLVLPTDGPTL LIHSGMTGRPYYCADGAAEDRYQRLVVSLDQGELRYTDLRKLRGVWLADDPDDLVPITGRQGPDSLGLGL RDFRDALTARSARRRQLKSALMEQSVLAGLGNLLVDEICWRARIRPTRAVADLDDDEVKALHRAMTQVLR TAVRHGRVPGLPRWLTGARDAPDPHCPRCGGRLDHARVGGRTTLWCPRCQPG
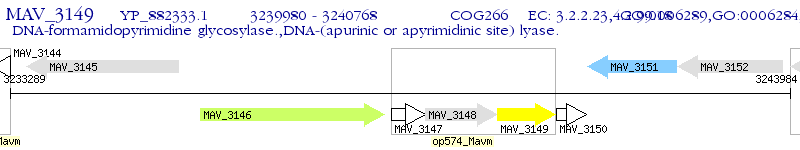
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3149 | - | - | 100% (262) | formamidopyrimidine DNA-glyxosylase |
| M. avium 104 | MAV_3782 | mutM | 6e-27 | 31.93% (285) | formamidopyrimidine-DNA glycosylase |
| M. avium 104 | MAV_1066 | - | 6e-21 | 29.39% (279) | formamidopyrimidine-DNA glycosylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2949c | fpg | 9e-27 | 33.68% (291) | formamidopyrimidine-DNA glycosylase |
| M. gilvum PYR-GCK | Mflv_4191 | - | 8e-28 | 32.04% (284) | formamidopyrimidine-DNA glycosylase |
| M. tuberculosis H37Rv | Rv2924c | fpg | 9e-27 | 33.68% (291) | formamidopyrimidine-DNA glycosylase |
| M. leprae Br4923 | MLBr_01658 | fpg | 5e-27 | 31.58% (285) | formamidopyrimidine-DNA glycosylase |
| M. abscessus ATCC 19977 | MAB_3255c | - | 1e-21 | 27.62% (286) | formamidopyrimidine-DNA glycosylase (MutM) |
| M. marinum M | MMAR_1783 | fpg | 5e-29 | 31.97% (294) | formamidopyrimidine-DNA glycosylase Fpg |
| M. smegmatis MC2 155 | MSMEG_2419 | mutM | 3e-26 | 31.23% (285) | formamidopyrimidine-DNA glycosylase |
| M. thermoresistible (build 8) | TH_1223 | fpg | 2e-28 | 31.93% (285) | PROBABLE FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE FPG (FAPY-DNA |
| M. ulcerans Agy99 | MUL_2031 | fpg | 4e-29 | 31.97% (294) | formamidopyrimidine-DNA glycosylase |
| M. vanbaalenii PYR-1 | Mvan_2172 | - | 7e-25 | 30.85% (295) | formamidopyrimidine-DNA glycosylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4191|M.gilvum_PYR-GCK MPELPEVEVVRRGLAAHVTGRTISAVRVHHPRAVRRHEAGPADLTARLLD
Mvan_2172|M.vanbaalenii_PYR-1 MPELPEVEVVRRGLAEHVTGRTVTAVRVHHPRAVRRHEAGPADLTARLLD
MSMEG_2419|M.smegmatis_MC2_155 MPELPEVEVVRRGLAEHVTGKTITGVRVHHPRAVRRHEAGPADLTARLLD
TH_1223|M.thermoresistible__bu VPELPEVEVVRRGLHEHVTGRAVAAVRVHHPRAVRRHEAGPADLTARLVD
Mb2949c|M.bovis_AF2122/97 MPELPEVEVVRRGLQAHVTGRTITEVRVHHPRAVRRHDAGPADLTARLRG
Rv2924c|M.tuberculosis_H37Rv MPELPEVEVVRRGLQAHVTGRTITEVRVHHPRAVRRHDAGPADLTARLRG
MMAR_1783|M.marinum_M MPELPEVEVVRRGLQDHVVGKTMTAVRVHHPRAVRRHEAGPADLTARLLG
MUL_2031|M.ulcerans_Agy99 MPELPEVEVVRRGLQDHVVGKTMTAVRVHHPRAVRRHEAGPADLTARLLG
MLBr_01658|M.leprae_Br4923 MPELPEVEVVRRGLQDYIVGKTITAVRVHHPRAVRRHVAGPTDLTNRLLG
MAB_3255c|M.abscessus_ATCC_199 MPELPEVEVVRRGLHHHLVGKTIASTLVYHERAVRRQSGGAVELAGLLAG
MAV_3149|M.avium_104 MPELPDVEGFRRQLADALPGRRVRRVKVHDPGILRN--TTATTLARRLTG
:****:** .** * : *: : . *:. :*. .. *: * .
Mflv_4191|M.gilvum_PYR-GCK SVITGTGRRGKYLWLTLGDGS-----------AVVVHLGMSG--QMLLGP
Mvan_2172|M.vanbaalenii_PYR-1 TTITGTGRRGKYLWLTLGDGADEPLARRESNFALVVHLGMSG--QMLLGD
MSMEG_2419|M.smegmatis_MC2_155 VRITGTGRRGKYLWLTLDDSA-----------ALVVHLGMSG--QMLLGP
TH_1223|M.thermoresistible__bu ARITGTGRRGKYLWLTLDDGA----------EALVVHLGMSG--QMLLGP
Mb2949c|M.bovis_AF2122/97 ARINGTDRRGKYLWLTLNTAG----VHRPTDTALVVHLGMSG--QMLLGA
Rv2924c|M.tuberculosis_H37Rv ARINGTDRRGKYLWLTLNTAG----VHRPTDTALVVHLGMSG--QMLLGA
MMAR_1783|M.marinum_M ARISGTDRRGKYLWLTLDAGAPTT-GRDDPDTALVVHLGMSG--QMLLGG
MUL_2031|M.ulcerans_Agy99 ARISGTDRRGKYLWLTLDAGAPTT-GRDDPDAALVVHLGMSG--QMLLGG
MLBr_01658|M.leprae_Br4923 TRINGIDRRGKYLWFLLDT-----------DIALVVHLGMSG--QMLLGT
MAB_3255c|M.abscessus_ATCC_199 QQISGTGRRGKYLWLTLEGSS--------GAQALVVHLGMSG--QMLIGP
MAV_3149|M.avium_104 RRFAGPRRHGKWLVLPTDGPT------------LLIHSGMTGRPYYCADG
: * *:**:* : :::* **:* .
Mflv_4191|M.gilvum_PYR-GCK VRNENHLRIAVLLDDGTALSFVDQRTFGGWMLAD----LVTVDGSDVPAP
Mvan_2172|M.vanbaalenii_PYR-1 VPNANHLRIAALLDDGTTLSFVDQRTFGGWMLAD----LVTVDGSDVPAP
MSMEG_2419|M.smegmatis_MC2_155 IRDTRHLRIAAVLDDGTALSFVDQRTFGGWQLTE----MVTVDGTDVPEP
TH_1223|M.thermoresistible__bu VPNENHLRIAALLDDGTTLSFVDQRTFGGWMIAD----MVTVEGSEVPAP
Mb2949c|M.bovis_AF2122/97 VPCAAHVRISALLDDGTVLSFADQRTFGGWLLAD----LVTVDGSVVPVP
Rv2924c|M.tuberculosis_H37Rv VPCAAHVRISALLDDGTVLSFADQRTFGGWLLAD----LVTVDGSVVPVP
MMAR_1783|M.marinum_M VPRAEHVRISAVLDDGTVLSFADQRTFGGWQLAD----LVSVDGSVVPAP
MUL_2031|M.ulcerans_Agy99 VPRAEHVRISAVLDDGTVLSFADQRTFGGWQLAD----LVSVDGSVVPAP
MLBr_01658|M.leprae_Br4923 VPRVDHVRISALFDDGTVLNFTDQRTLGGWLLAD----LMTVDGSVLPVP
MAB_3255c|M.abscessus_ATCC_199 ISRPQHLRIAATLDDGSVLSFVDQRTFGGWMVTD----LVTVDGSELPEP
MAV_3149|M.avium_104 AAEDRYQRLVVSLDQG-ELRYTDLRKLRGVWLADDPDDLVPITGRQGPDS
: *: . :*:* * :.* *.: * ::: ::.: * * .
Mflv_4191|M.gilvum_PYR-GCK VAHIARDPLDPLFDRAAVVNVLRRKHSEIKRQLLDQTVVSGIGNIYADES
Mvan_2172|M.vanbaalenii_PYR-1 VAHIARDPLDPLFDRDAVVKVLRRKHSEIKRQLLDQTVVSGIGNIYADES
MSMEG_2419|M.smegmatis_MC2_155 VAHIARDPLDPLFDRDRVVTVLRGKHSEIKRQLLDQTVVSGIGNIYADEA
TH_1223|M.thermoresistible__bu VVHLARDPLDPLFDRDAVVTVLRGKHSEIKRQLLDQTVVSGIGNIYADEA
Mb2949c|M.bovis_AF2122/97 VAHLARDPLDPRFDCDAVVKVLRRKHSELKRQLLDQRVVSGIGNIYADEA
Rv2924c|M.tuberculosis_H37Rv VAHLARDPLDPRFDCDAVVKVLRRKHSELKRQLLDQRVVSGIGNIYADEA
MMAR_1783|M.marinum_M VAHLARDPLDPLFDVDSVIKVLRGKHSEIKRQLLDQQVVSGIGNIYADEA
MUL_2031|M.ulcerans_Agy99 VAHLARDPLDPLFDVDSVIKVLRGKHSEVKRRLLDQQVVSGIGNIYADEA
MLBr_01658|M.leprae_Br4923 VAHLARDPFDPRFDVEAVVKVLRCKHSELKRQLLDQQTVSGIGNIYADEA
MAB_3255c|M.abscessus_ATCC_199 VAHIARDPLDELFEIGAVVTRLRGKHTEIKRALLDQTVVSGVGNIYADEA
MAV_3149|M.avium_104 LGLGLRDFRDALTARS-------ARRRQLKSALMEQSVLAGLGNLLVDEI
: ** * :: ::* *::* .::*:**: .**
Mflv_4191|M.gilvum_PYR-GCK LWRAKINGARLASGVSKAKLAELLDAATDVMTDALAQGGTSFDSLYVNVN
Mvan_2172|M.vanbaalenii_PYR-1 LWRAKINGARLASGVSRAKLAELLGAAADVMTDALAQGGTSFDSLYVNVN
MSMEG_2419|M.smegmatis_MC2_155 LWRTKINGARIAAALPRRRLAELLDAAAEVMTDALGQGGTSFDSLYVNVN
TH_1223|M.thermoresistible__bu LWRARINGARTASALTRRRLGELLDAAADVMREALRQGGTSFDSLYVNVN
Mb2949c|M.bovis_AF2122/97 LWRAKVNGAHVAATLRCRRLGAVLHAAADVMREALAKGGTSFDSLYVNVN
Rv2924c|M.tuberculosis_H37Rv LWRAKVNGAHVAATLRCRRLGAVLHAAADVMREALAKGGTSFDSLYVNVN
MMAR_1783|M.marinum_M LWRAKVHGARVAATLTRRQLGAVLDAAADVMREALAKGGTSFDSLYVNVN
MUL_2031|M.ulcerans_Agy99 LWRAKVHGARVAATLTRRQLGAVLDAAADVMREALAKGGTSFDSLYVNVN
MLBr_01658|M.leprae_Br4923 LWRAEVHGARIAATLTRRQLAAVLDAAADVMRDSLAKGGTSFDSLYVNVN
MAB_3255c|M.abscessus_ATCC_199 LWQARVHGRRLTDGMSRAKLTEVLDSAAAVMRLALAQGGTSFDDLYVNVN
MAV_3149|M.avium_104 CWRARIRPTRAVADLDDDEVKALHRAMTQVLRTAVRHG---------RVP
*::.:. : . : .: : : : *: :: :* .*
Mflv_4191|M.gilvum_PYR-GCK GESGYFDRSLDAYGREGEPCRRCGAIMRRDKFMNRSSFYCPRCQ--PRPR
Mvan_2172|M.vanbaalenii_PYR-1 GESGYFDRSLDAYGREGEPCRRCGAIMRRDKFMNRSSFYCPRCQ--PRPR
MSMEG_2419|M.smegmatis_MC2_155 GESGYFDRSLDAYGREGEPCRRCGAIMRREKFMNRSSFYCPRCQ--PRPR
TH_1223|M.thermoresistible__bu GESGYFERSLDAYGRAGEPCRRCGAVLRREKFMNRSSFYCPKCQ--PRPR
Mb2949c|M.bovis_AF2122/97 GESGYFERSLDAYGREGENCRRCGAVIRRERFMNRSSFYCPRCQ--PRPR
Rv2924c|M.tuberculosis_H37Rv GESGYFERSLDAYGREGENCRRCGAVIRRERFMNRSSFYCPRCQ--PRPR
MMAR_1783|M.marinum_M GQSGYFDRSLDAYGREGEHCRRCGAVMRREKFMNRSSFYCPRCQ--PRPR
MUL_2031|M.ulcerans_Agy99 GQSGYFDRSLDAYGREGEHCRRCGAVMRREKFMNRSSFYCPRCQ--PRPR
MLBr_01658|M.leprae_Br4923 GESGYFDRSLDAYGREGEGCRRCGAVMHREKFMNRSSFYCPRCQ--PRPR
MAB_3255c|M.abscessus_ATCC_199 GESGYFDRSLEAYGREGEPCRRCGRAMRREAFMNRSSYFCPSCQRLVEPR
MAV_3149|M.avium_104 GLPRWLTGARDAP---DPHCPRCGGRLDHARVGGRTTLWCPRCQ----PG
* . :: : :* . * *** : : . .*:: :** ** *
Mflv_4191|M.gilvum_PYR-GCK V---
Mvan_2172|M.vanbaalenii_PYR-1 V---
MSMEG_2419|M.smegmatis_MC2_155 VPRV
TH_1223|M.thermoresistible__bu IRRC
Mb2949c|M.bovis_AF2122/97 K---
Rv2924c|M.tuberculosis_H37Rv K---
MMAR_1783|M.marinum_M R---
MUL_2031|M.ulcerans_Agy99 R---
MLBr_01658|M.leprae_Br4923 R---
MAB_3255c|M.abscessus_ATCC_199 ----
MAV_3149|M.avium_104 ----