For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PELPEVEVVRRGLAAHVTGRTISAVRVHHPRAVRRHEAGPADLTARLLDSVITGTGRRGKYLWLTLGDGS AVVVHLGMSGQMLLGPVRNENHLRIAVLLDDGTALSFVDQRTFGGWMLADLVTVDGSDVPAPVAHIARDP LDPLFDRAAVVNVLRRKHSEIKRQLLDQTVVSGIGNIYADESLWRAKINGARLASGVSKAKLAELLDAAT DVMTDALAQGGTSFDSLYVNVNGESGYFDRSLDAYGREGEPCRRCGAIMRRDKFMNRSSFYCPRCQPRPR V*
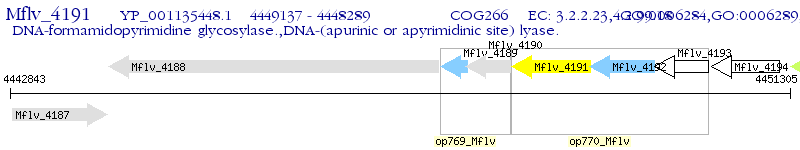
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4191 | - | - | 100% (282) | formamidopyrimidine-DNA glycosylase |
| M. gilvum PYR-GCK | Mflv_1852 | - | 1e-20 | 28.14% (295) | DNA-formamidopyrimidine glycosylase |
| M. gilvum PYR-GCK | Mflv_4811 | - | 4e-16 | 27.46% (284) | formamidopyrimidine-DNA glycolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2949c | fpg | 1e-126 | 76.74% (288) | formamidopyrimidine-DNA glycosylase |
| M. tuberculosis H37Rv | Rv2924c | fpg | 1e-126 | 76.74% (288) | formamidopyrimidine-DNA glycosylase |
| M. leprae Br4923 | MLBr_01658 | fpg | 1e-122 | 74.02% (281) | formamidopyrimidine-DNA glycosylase |
| M. abscessus ATCC 19977 | MAB_3255c | - | 1e-112 | 70.00% (280) | formamidopyrimidine-DNA glycosylase (MutM) |
| M. marinum M | MMAR_1783 | fpg | 1e-127 | 76.29% (291) | formamidopyrimidine-DNA glycosylase Fpg |
| M. avium 104 | MAV_3782 | mutM | 1e-130 | 79.86% (283) | formamidopyrimidine-DNA glycosylase |
| M. smegmatis MC2 155 | MSMEG_2419 | mutM | 1e-142 | 85.82% (282) | formamidopyrimidine-DNA glycosylase |
| M. thermoresistible (build 8) | TH_1223 | fpg | 1e-140 | 83.75% (283) | PROBABLE FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE FPG (FAPY-DNA |
| M. ulcerans Agy99 | MUL_2031 | fpg | 1e-127 | 75.60% (291) | formamidopyrimidine-DNA glycosylase |
| M. vanbaalenii PYR-1 | Mvan_2172 | - | 1e-151 | 90.44% (293) | formamidopyrimidine-DNA glycosylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4191|M.gilvum_PYR-GCK MPELPEVEVVRRGLAAHVTGRTISAVRVHHPRAVRRHEAGPADLTARLLD
Mvan_2172|M.vanbaalenii_PYR-1 MPELPEVEVVRRGLAEHVTGRTVTAVRVHHPRAVRRHEAGPADLTARLLD
MSMEG_2419|M.smegmatis_MC2_155 MPELPEVEVVRRGLAEHVTGKTITGVRVHHPRAVRRHEAGPADLTARLLD
TH_1223|M.thermoresistible__bu VPELPEVEVVRRGLHEHVTGRAVAAVRVHHPRAVRRHEAGPADLTARLVD
Mb2949c|M.bovis_AF2122/97 MPELPEVEVVRRGLQAHVTGRTITEVRVHHPRAVRRHDAGPADLTARLRG
Rv2924c|M.tuberculosis_H37Rv MPELPEVEVVRRGLQAHVTGRTITEVRVHHPRAVRRHDAGPADLTARLRG
MLBr_01658|M.leprae_Br4923 MPELPEVEVVRRGLQDYIVGKTITAVRVHHPRAVRRHVAGPTDLTNRLLG
MMAR_1783|M.marinum_M MPELPEVEVVRRGLQDHVVGKTMTAVRVHHPRAVRRHEAGPADLTARLLG
MUL_2031|M.ulcerans_Agy99 MPELPEVEVVRRGLQDHVVGKTMTAVRVHHPRAVRRHEAGPADLTARLLG
MAV_3782|M.avium_104 MPELPEVEVVRRGLHSHVVGKTIGAVRVHHPRAVRRHEAGPADLTARLLG
MAB_3255c|M.abscessus_ATCC_199 MPELPEVEVVRRGLHHHLVGKTIASTLVYHERAVRRQSGGAVELAGLLAG
:************* ::.*::: . *:* *****: .*..:*: * .
Mflv_4191|M.gilvum_PYR-GCK SVITGTGRRGKYLWLTLGDGS-----------AVVVHLGMSGQMLLGPVR
Mvan_2172|M.vanbaalenii_PYR-1 TTITGTGRRGKYLWLTLGDGADEPLARRESNFALVVHLGMSGQMLLGDVP
MSMEG_2419|M.smegmatis_MC2_155 VRITGTGRRGKYLWLTLDDSA-----------ALVVHLGMSGQMLLGPIR
TH_1223|M.thermoresistible__bu ARITGTGRRGKYLWLTLDDGA----------EALVVHLGMSGQMLLGPVP
Mb2949c|M.bovis_AF2122/97 ARINGTDRRGKYLWLTLNTAGVHR----PTDTALVVHLGMSGQMLLGAVP
Rv2924c|M.tuberculosis_H37Rv ARINGTDRRGKYLWLTLNTAGVHR----PTDTALVVHLGMSGQMLLGAVP
MLBr_01658|M.leprae_Br4923 TRINGIDRRGKYLWFLLD-----------TDIALVVHLGMSGQMLLGTVP
MMAR_1783|M.marinum_M ARISGTDRRGKYLWLTLDAGAPTTG-RDDPDTALVVHLGMSGQMLLGGVP
MUL_2031|M.ulcerans_Agy99 ARISGTDRRGKYLWLTLDAGAPTTG-RDDPDAALVVHLGMSGQMLLGGVP
MAV_3782|M.avium_104 ARITGTDRRGKYLWLLLDG----------RDTALVVHLGMSGQMLLGAVP
MAB_3255c|M.abscessus_ATCC_199 QQISGTGRRGKYLWLTLEGSS--------GAQALVVHLGMSGQMLIGPIS
*.* .*******: * *:***********:* :
Mflv_4191|M.gilvum_PYR-GCK NENHLRIAVLLDDGTALSFVDQRTFGGWMLADLVTVDGSDVPAPVAHIAR
Mvan_2172|M.vanbaalenii_PYR-1 NANHLRIAALLDDGTTLSFVDQRTFGGWMLADLVTVDGSDVPAPVAHIAR
MSMEG_2419|M.smegmatis_MC2_155 DTRHLRIAAVLDDGTALSFVDQRTFGGWQLTEMVTVDGTDVPEPVAHIAR
TH_1223|M.thermoresistible__bu NENHLRIAALLDDGTTLSFVDQRTFGGWMIADMVTVEGSEVPAPVVHLAR
Mb2949c|M.bovis_AF2122/97 CAAHVRISALLDDGTVLSFADQRTFGGWLLADLVTVDGSVVPVPVAHLAR
Rv2924c|M.tuberculosis_H37Rv CAAHVRISALLDDGTVLSFADQRTFGGWLLADLVTVDGSVVPVPVAHLAR
MLBr_01658|M.leprae_Br4923 RVDHVRISALFDDGTVLNFTDQRTLGGWLLADLMTVDGSVLPVPVAHLAR
MMAR_1783|M.marinum_M RAEHVRISAVLDDGTVLSFADQRTFGGWQLADLVSVDGSVVPAPVAHLAR
MUL_2031|M.ulcerans_Agy99 RAEHVRISAVLDDGTVLSFADQRTFGGWQLADLVSVDGSVVPAPVAHLAR
MAV_3782|M.avium_104 RAEHVRISALLDDGTVLSFADQRTFGGWMLADLLEVDGSVVPEPVAHLAR
MAB_3255c|M.abscessus_ATCC_199 RPQHLRIAATLDDGSVLSFVDQRTFGGWMVTDLVTVDGSELPEPVAHIAR
*:**:. :***:.*.*.****:*** ::::: *:*: :* **.*:**
Mflv_4191|M.gilvum_PYR-GCK DPLDPLFDRAAVVNVLRRKHSEIKRQLLDQTVVSGIGNIYADESLWRAKI
Mvan_2172|M.vanbaalenii_PYR-1 DPLDPLFDRDAVVKVLRRKHSEIKRQLLDQTVVSGIGNIYADESLWRAKI
MSMEG_2419|M.smegmatis_MC2_155 DPLDPLFDRDRVVTVLRGKHSEIKRQLLDQTVVSGIGNIYADEALWRTKI
TH_1223|M.thermoresistible__bu DPLDPLFDRDAVVTVLRGKHSEIKRQLLDQTVVSGIGNIYADEALWRARI
Mb2949c|M.bovis_AF2122/97 DPLDPRFDCDAVVKVLRRKHSELKRQLLDQRVVSGIGNIYADEALWRAKV
Rv2924c|M.tuberculosis_H37Rv DPLDPRFDCDAVVKVLRRKHSELKRQLLDQRVVSGIGNIYADEALWRAKV
MLBr_01658|M.leprae_Br4923 DPFDPRFDVEAVVKVLRCKHSELKRQLLDQQTVSGIGNIYADEALWRAEV
MMAR_1783|M.marinum_M DPLDPLFDVDSVIKVLRGKHSEIKRQLLDQQVVSGIGNIYADEALWRAKV
MUL_2031|M.ulcerans_Agy99 DPLDPLFDVDSVIKVLRGKHSEVKRRLLDQQVVSGIGNIYADEALWRAKV
MAV_3782|M.avium_104 DPLDPRFDADAVVKVLRRKHSEIKRQLLDQQVVSGIGNIYADEALWRAKV
MAB_3255c|M.abscessus_ATCC_199 DPLDELFEIGAVVTRLRGKHTEIKRALLDQTVVSGVGNIYADEALWQARV
**:* *: *:. ** **:*:** **** .***:*******:**::.:
Mflv_4191|M.gilvum_PYR-GCK NGARLASGVSKAKLAELLDAATDVMTDALAQGGTSFDSLYVNVNGESGYF
Mvan_2172|M.vanbaalenii_PYR-1 NGARLASGVSRAKLAELLGAAADVMTDALAQGGTSFDSLYVNVNGESGYF
MSMEG_2419|M.smegmatis_MC2_155 NGARIAAALPRRRLAELLDAAAEVMTDALGQGGTSFDSLYVNVNGESGYF
TH_1223|M.thermoresistible__bu NGARTASALTRRRLGELLDAAADVMREALRQGGTSFDSLYVNVNGESGYF
Mb2949c|M.bovis_AF2122/97 NGAHVAATLRCRRLGAVLHAAADVMREALAKGGTSFDSLYVNVNGESGYF
Rv2924c|M.tuberculosis_H37Rv NGAHVAATLRCRRLGAVLHAAADVMREALAKGGTSFDSLYVNVNGESGYF
MLBr_01658|M.leprae_Br4923 HGARIAATLTRRQLAAVLDAAADVMRDSLAKGGTSFDSLYVNVNGESGYF
MMAR_1783|M.marinum_M HGARVAATLTRRQLGAVLDAAADVMREALAKGGTSFDSLYVNVNGQSGYF
MUL_2031|M.ulcerans_Agy99 HGARVAATLTRRQLGAVLDAAADVMREALAKGGTSFDSLYVNVNGQSGYF
MAV_3782|M.avium_104 HGARIADALTRKQLTAVLDAAADVMRDALAKGGTSFDSLYVNVNGESGYF
MAB_3255c|M.abscessus_ATCC_199 HGRRLTDGMSRAKLTEVLDSAAAVMRLALAQGGTSFDDLYVNVNGESGYF
:* : : : :* :* :*: ** :* :******.*******:****
Mflv_4191|M.gilvum_PYR-GCK DRSLDAYGREGEPCRRCGAIMRRDKFMNRSSFYCPRCQ--PRPRV---
Mvan_2172|M.vanbaalenii_PYR-1 DRSLDAYGREGEPCRRCGAIMRRDKFMNRSSFYCPRCQ--PRPRV---
MSMEG_2419|M.smegmatis_MC2_155 DRSLDAYGREGEPCRRCGAIMRREKFMNRSSFYCPRCQ--PRPRVPRV
TH_1223|M.thermoresistible__bu ERSLDAYGRAGEPCRRCGAVLRREKFMNRSSFYCPKCQ--PRPRIRRC
Mb2949c|M.bovis_AF2122/97 ERSLDAYGREGENCRRCGAVIRRERFMNRSSFYCPRCQ--PRPRK---
Rv2924c|M.tuberculosis_H37Rv ERSLDAYGREGENCRRCGAVIRRERFMNRSSFYCPRCQ--PRPRK---
MLBr_01658|M.leprae_Br4923 DRSLDAYGREGEGCRRCGAVMHREKFMNRSSFYCPRCQ--PRPRR---
MMAR_1783|M.marinum_M DRSLDAYGREGEHCRRCGAVMRREKFMNRSSFYCPRCQ--PRPRR---
MUL_2031|M.ulcerans_Agy99 DRSLDAYGREGEHCRRCGAVMRREKFMNRSSFYCPRCQ--PRPRR---
MAV_3782|M.avium_104 DRSLDAYGREGESCRRCGAVMRREKFMNRSSFYCPKCQ--PRPRL---
MAB_3255c|M.abscessus_ATCC_199 DRSLEAYGREGEPCRRCGRAMRREAFMNRSSYFCPSCQRLVEPR----
:***:**** ** ***** ::*: ******::** ** .**