For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTRYEEPNRAARAANVVVRWLADAGVSIAGTRALRVRGRKTGKLRAVVVNLLTVDGTDYLVSPRGDTQW ARNVRAASVVELGPRWRRRRMRISEVDDAAKPQLLRRYLDRWYWQVKDYVAGLTPDSSDEQFRAAAPTIP VFALTKAG
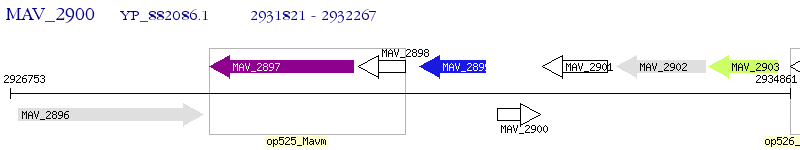
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2900 | - | - | 100% (148) | hypothetical protein MAV_2900 |
| M. avium 104 | MAV_4595 | - | 1e-08 | 31.69% (142) | cell entry (mce) related family protein |
| M. avium 104 | MAV_0161 | - | 5e-07 | 30.28% (142) | hypothetical protein MAV_0161 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3415 | - | 6e-46 | 59.03% (144) | hypothetical protein Mflv_3415 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3001 | - | 1e-09 | 33.33% (141) | hypothetical protein MAB_3001 |
| M. marinum M | MMAR_2916 | - | 0.0001 | 25.90% (139) | hypothetical protein MMAR_2916 |
| M. smegmatis MC2 155 | MSMEG_3660 | - | 2e-54 | 68.06% (144) | hypothetical protein MSMEG_3660 |
| M. thermoresistible (build 8) | TH_4256 | - | 2e-08 | 35.65% (115) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2188 | - | 7e-05 | 25.90% (139) | hypothetical protein MUL_2188 |
| M. vanbaalenii PYR-1 | Mvan_3168 | - | 2e-52 | 66.67% (144) | hypothetical protein Mvan_3168 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3415|M.gilvum_PYR-GCK -MNERYDAPGPAARIFNGAIRRLAESGISIQGSTAVRVRGRSTGRLRGVV
Mvan_3168|M.vanbaalenii_PYR-1 -MSTRYDAPGTGARIFNDVVRWCADVGVSIQGSTAIRVRGRTTGRLRGVV
MSMEG_3660|M.smegmatis_MC2_155 --MTRYDEPGPVARAANTLIRWLAEHGVSIAGSTALRVRGRRSGKLRSVV
MAV_2900|M.avium_104 -MSTRYEEPNRAARAANVVVRWLADAGVSIAGTRALRVRGRKTGKLRAVV
MMAR_2916|M.marinum_M ---MSNPKPPAWLKPMNKAMVTLQKIGVVAGPVRVLTVTGRKTGLPRSTP
MUL_2188|M.ulcerans_Agy99 ---MSNPKPPAWLKPMNKAMVTLQKIGVVAGPVRVLTVTGRKTGLPRSTP
MAB_3001|M.abscessus_ATCC_1997 MTTVTNTQPPRWLKPMNKVVRAFHRLGVSTGPAMVLTVPGRKTGQPRPTP
TH_4256|M.thermoresistible__bu ---MAYLKPP----WFTRAVFNKIAMALGAGGSEILTVTRRVSRQPQQIP
* . : .: : * * : :
Mflv_3415|M.gilvum_PYR-GCK VNLLTVDGRRYLVSPRGNTQWARNARAAGEIEIGPRRRSRIHPVVELSDD
Mvan_3168|M.vanbaalenii_PYR-1 VNLLAVDGRRYLVSPRGNTQWARNARAAGQVEVGPRWRSRPHTVVEIADD
MSMEG_3660|M.smegmatis_MC2_155 VNLMTVDGHRYVVAPRGETEWVRNARAAGVVEIGPRRRRRTAQIAEIPDD
MAV_2900|M.avium_104 VNLLTVDGTDYLVSPRGDTQWARNVRAASVVELGPRWRRRRMRISEVDDA
MMAR_2916|M.marinum_M ATPFVLDDCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVVFTELTAA
MUL_2188|M.ulcerans_Agy99 ATPFVLDDCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVVFTELTAA
MAB_3001|M.abscessus_ATCC_1997 ITPFEVDGQLFAVGGYPGSDWARNAAAAGSGTLTKGNKVRHIRIVTVPPH
TH_4256|M.thermoresistible__bu VIPVDVDGNRHLVSTRGETEWVRNVRAEPNVLLGDTR----MVAREVPPE
. :*. . *. ::*.**. * : :
Mflv_3415|M.gilvum_PYR-GCK TKPAVLKPYVDRWFWQVKGHVG-GLTPQSGRDELRAAAPSIPVFEILN--
Mvan_3168|M.vanbaalenii_PYR-1 AKPELLRRYLDRWYWQVKGHVG-GLTPESTPAEIREVAPSIPVFELG---
MSMEG_3660|M.smegmatis_MC2_155 AKPTLLKRYLDRWYWEVKGHMA-DLTPKSSAEQLRAAAPHIPVFALVETS
MAV_2900|M.avium_104 AKPQLLRRYLDRWYWQVKDYVA-GLTPDSSDEQFRAAAPTIPVFALTKAG
MMAR_2916|M.marinum_M QAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAVFRLDPTA
MUL_2188|M.ulcerans_Agy99 QAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAVFRLDPTA
MAB_3001|M.abscessus_ATCC_1997 VARRVLREFALQVPVGVRFAKSAGLVRQGTPDEFEGLAGTLSVFRFDPD-
TH_4256|M.thermoresistible__bu SRAPILAAYRQKAGRVVKGYFE--ALPDDADHPVFLLTPVSKA-------
:* : : * .. . : .
Mflv_3415|M.gilvum_PYR-GCK --
Mvan_3168|M.vanbaalenii_PYR-1 --
MSMEG_3660|M.smegmatis_MC2_155 --
MAV_2900|M.avium_104 --
MMAR_2916|M.marinum_M QN
MUL_2188|M.ulcerans_Agy99 QN
MAB_3001|M.abscessus_ATCC_1997 --
TH_4256|M.thermoresistible__bu --